Figure 5.
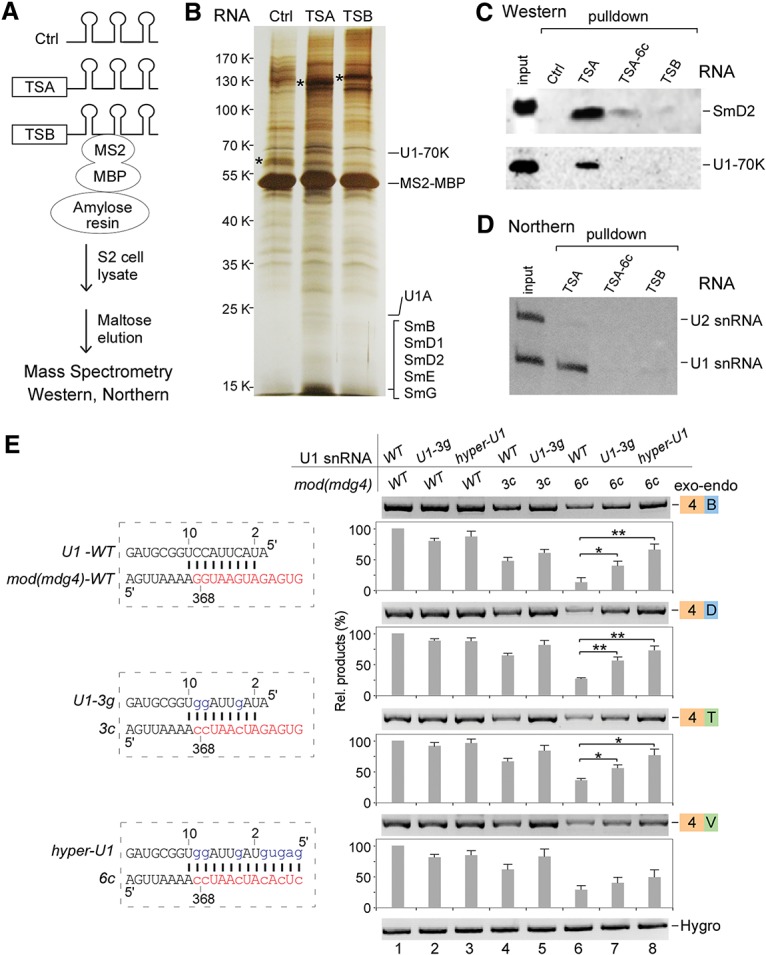
The TSA core motif binds with U1 snRNP to facilitate trans-splicing of mod(mdg4). (A) Schematic of RNA affinity purification. (Ctrl) 6xMS2-RNA-binding sites only. (B) Silver staining of affinity-purified proteins. Analyzed by mass spectrometry, TSA and TSB specifically associated proteins are listed in Supplemental Table S1. Proteins that specifically associated with TSA are indicated. (*) Visualized RNAs. (C) Western blot analyses of TSA RNA-associated U1 snRNP proteins. (TSA-6c) 6Gs-to-6Cs mutations in the TSA core motif. (D) Northern blot analysis revealed that U1 snRNA is specifically associated with TSA. (E) Compensatory changes in U1 snRNA partially rescue the decreased trans-splicing activities of the TSA core motif mutations. The wild-type (WT) TSA core motif forms 9 base pairs (bp) with the 5′ end of U1 snRNA. mod(mdg4) mutants 3c and 6c disrupt this base-pairing, but U1-3g and hyper-U1 mutants will restore or enhance the base-pairing. Relative trans-spliced products were normalized to loading controls and the wild-type constructs. Mean ± SEM; n = 3; (*) P < 0.05; (**) P < 0.01.
