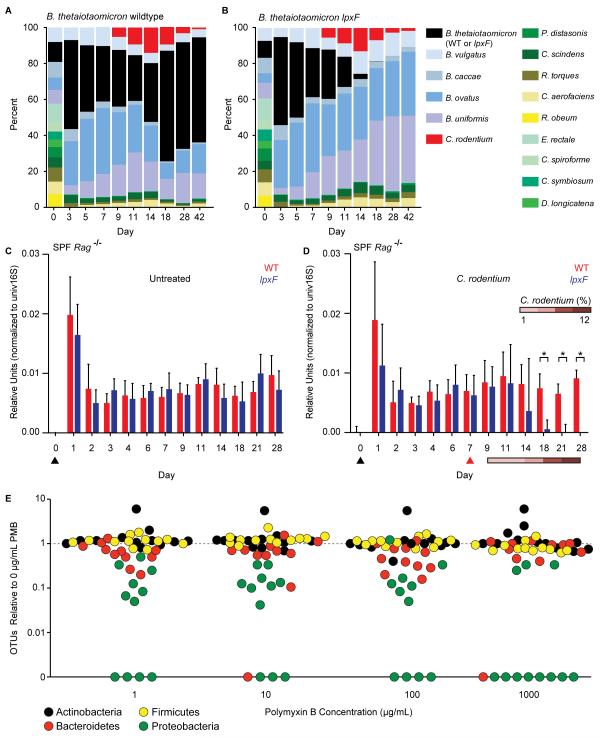
Fig. 4. AMP resistance determines commensal resilience in the context of a human or murine gut microbiota.
(A, B) Germfree mice (n=5/group) were colonized with 14 prominent human gut microbes, including B. thetaiotaomicron wildtype or lpxF deletion mutant, 7 days prior to infection with C. rodentium; community composition was monitored by species-specific quantitative polymerase chain reaction (qPCR) and reported as median percent of total. Uninfected controls are shown in fig. S6B. (C, D) Specific pathogen free Rag−/− mice (n=5/group) were gavaged (black arrowhead) with either B. thetaiotaomicron wildtype or lpxF deletion mutant; in (D), mice were infected with C. rodentium 7 days later (red arrowhead). Colonization levels were assessed by qPCR from fecal DNA. C. rodentium levels are reported percentage of total fecal DNA. Error bars indicate standard deviation and asterisks indicate significance (p<0.05). (E) AMP resistance is a general feature of human gut Bacteroidetes, Firmicutes, and Actinobacteria. Fecal samples from 12 unrelated, healthy human donors were cultured on varying PMB concentrations (x-axis) and the number of species-level phylotypes (OTUs) belonging to each phylum observed from each donor (points, colored by phylum) was normalized to the number observed in culture in the absence of PMB. A weighted (abundance) analysis provides similar results (fig. S8B).