Abstract
The unfolded protein response (UPR) has traditionally been viewed as an adaptive response triggered upon accumulation of unfolded proteins in the endoplasmic reticulum (ER), aimed at restoring ER function. The UPR can also be an anticipatory response that is activated well before the disruption of protein homeostasis. UPR signaling intersects at many levels with the innate and adaptive immune response. In some immune cell types like dendritic cells and B cells, particular UPR sensors appear constitutively active in the absence of traditional UPR gene program induction, necessary for antigen presentation and immunoglobulin synthesis. The UPR also influences Toll-like receptor signaling and NF-κB activation, and some pathogens subvert the UPR. This review summarizes these emerging non-canonical functions of the UPR in immunity.
The endoplasmic reticulum (ER) is the production and folding factory of secreted and transmembrane proteins of the cell. It is well adapted for this function and performs complex protein modifications, such as glycosylation and disulfide bond formation1. Quality control ensures that only properly folded proteins exit the ER via the secretory pathway, while improperly folded proteins exit the ER through ER-associated degradation (ERAD) or via autophagy2. Depending on the physiological and environmental demand, the protein production rate in the ER can increase very rapidly3. An imbalance between the folding load of nascent proteins entering the ER and the capacity of the ER to handle this load causes ER stress. This is detected by three sensors that face the ER lumen: inositol-requiring enzyme 1 (IRE1 also known as ERN1, ER to nucleus signaling 1), protein kinase R-like ER kinase (PERK) and activating transcription factor 6 (ATF6) (Fig. 1). They trigger the unfolded protein response (UPR), an adaptive response aimed at restoring protein-folding homeostasis by three main mechanisms: transient reduction in protein translation4, 5, 6; increase in the folding capacity and ERAD7; and initiation of programmed cell death, when ER stress cannot be resolved8.
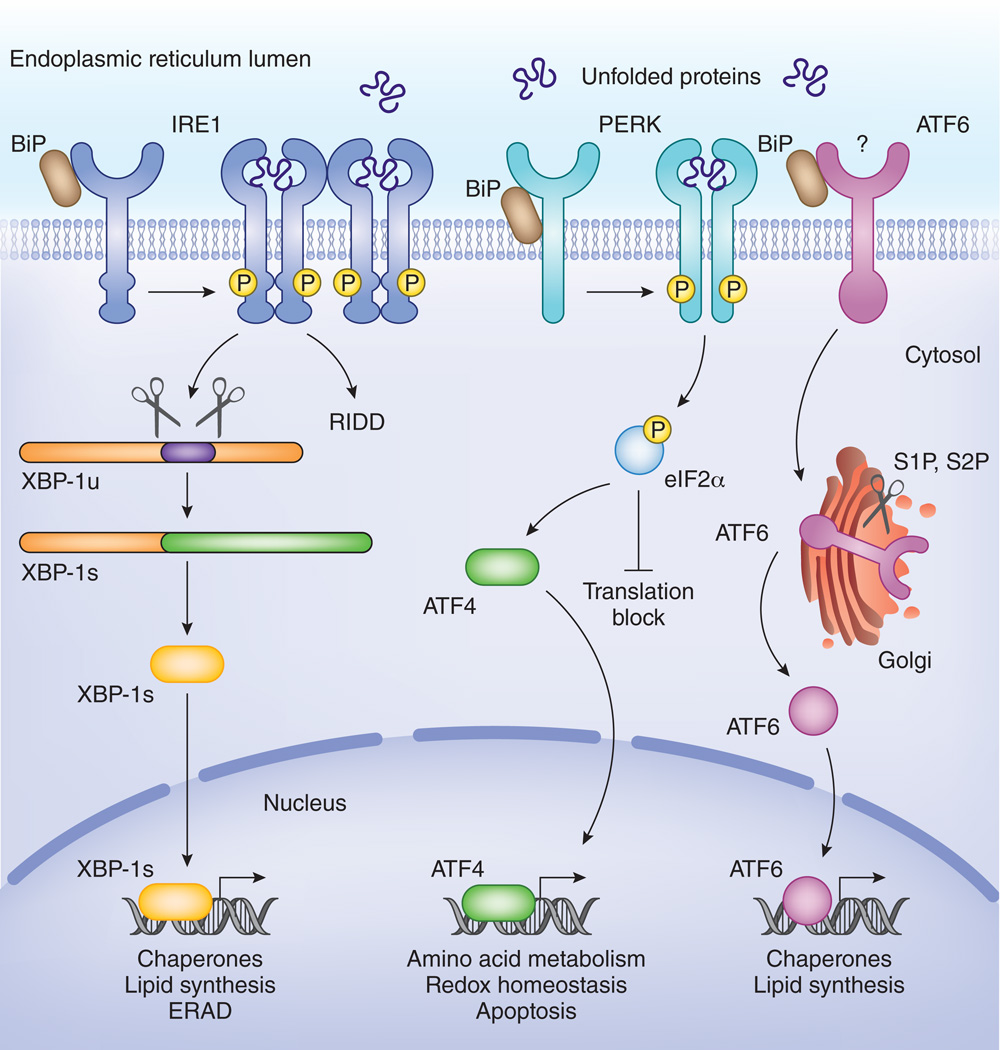
Figure 1. Three different sensors jointly coordinate the UPR in mammals.
IRE1 and PERK are activated by oligomerization and transphosphorylation upon binding of unfolded proteins and release of the chaperone BiP. The endonuclease domain of IRE1 cleaves XBP-1 mRNA in an unconventional splicing reaction to generate XBP-1s mRNA, and the resulting product encodes a transcription factor of the bZIP family. In addition the endonuclease activity of IRE-1 also contributes to Regulated IRE-1 dependent decay (RIDD), the degradation of mRNAs that are recruited at ribosomes at the ER. PERK is a transmembrane kinase that phosphorylates eukaryotic initiation factor 2 alpha (eIF2α). This leads to a transient inhibition of protein translation. Some mRNAs that encode small upstream open reading frames (uORFs) in their 5’ UTR escape the translation stop, the most prominent being the transcription factor ATF4. ATF4 in turn triggers expression of CHOP, GADD34 and additional factors important for amino acid metabolism and redox control. ATF6 is activated upon release of BiP and translocated to the Golgi where it undergoes sequential cleavage and removal of its luminal domain. The remaining transactivation domain of ATF6 moves to the nucleus and coordinates expression of genes involved in chaperone pathways or lipid biosynthesis.
IRE1α is a bifunctional type I transmembrane protein that harbors a serine/threonine kinase domain and a unique ribonuclease (RNAse) domain in its cytoplasmic domain. The luminal domain senses unfolded proteins and is responsible for high-order oligomerization of IRE1α. Oligomerization activates the RNase domain, which cleaves XBP1 (X-box binding protein) mRNA at two discrete stem loop structures through an unconventional cytoplasmic splicing reaction9, 10. The resulting fragments are ligated by an as yet unidentified – at least in mammals – RNA ligase, yielding an active transcription factor of the bZIP family, XBP-1s9, 10. XBP-1s plays a major role in the induction of genes involved in lipid biosynthesis, ERAD and chaperone production11, 12, 13, 14. In Saccharomyces cerevisiae, the XBP1 homolog HAC1 mRNA is the only known substrate of IRE1. In fission yeast (S. pombe) and metazoans however, the IRE1 has more promiscuous endonuclease activity, cleaving many mRNAs that are localized to the ER membrane6, 15. IRE1 dependent decay of mRNAs (termed RIDD) presumably represents another mechanism for the attenuation of protein translation5. Upon oligomerization, IRE1α autophosphorylates via the activity of its kinase domain. The physiological significance of this is unclear, as only nucleotide binding and not phosphotransfer appears to be required for RNase activity16, although phoshorylation may drive further oligomerization17. Alternatively, phosphorylated sites in IRE1α might serve as docking sites for TRAF2 and enable ER stress-induced JNK activation18. Mammals have a second isoform of IRE119. Unlike IRE1α, IRE1β does not cleave XBP1 mRNA and its expression is restricted to epithelial cells lining the gut and lung20, 21, 22, where it may control RIDD22, 23, 24.
The ATF6 sensor is a type II transmembrane protein with its carboxy-terminus facing the ER lumen and a bZIP transcription factor in its amino-terminus25, 26 (Fig. 1). In response to ER stress, ATF6 is transported via COPII vesicles to the Golgi apparatus where it undergoes regulated intramembrane proteolysis by sequential cleavage by site 1 and site 2 proteases (S1P and S2P) in a manner reminiscent of SREBP transcription factor activation27. This releases the amino-terminal transcription factor fragment pATF6-N that moves to the nucleus and directs expression of UPR genes involved in ER membrane expansion, the ERAD pathway and folding26, 28, 29. Several other bZIP transcription family members, such as OASIS or CREB3, are related to ATF6 and are also cleaved by regulated intramembrane proteolysis30. Some of them such as CREBH, do not trigger genes that enhance capacity of the ER but rather trigger acute phase proteins, representing an intriguing link between ER stress and inflammation31.
Finally, PERK, is another type I transmembrane protein kinase that shares about 20% similarity in its luminal domain with IRE14, 32. Unlike IRE1, PERK has a well-established cytoplasmic kinase substrate, the eukaryotic initiation factor 2 alpha (eIF2α)(Figure 1)33. Phosphorylation of eIF2α leads to an inhibition of the guanine exchange factor eIF2B that recycles eIF2 to its active GTP bound form. As such, there is a delay in ternary complex formation and a strong reduction in cap-dependent translation, which is thought to be essential for cell survival in conditions of ER stress34, 35. A few transcripts with short upstream open reading frames (uORFs) in their 5’ UTR escape inhibition of translation under conditions of high eIF2a phosphorylation and mediate a PERK-dependent transcriptional response33. ATF4 is a bZIP transcription factor induced in this manner. ATF4 directs the expression of CHOP, a proapoptotic factor33 and GADD34, a negative feedback regulator of eIF2α phosphorylation36. Additionally, ATF4 coordinates a gene program needed for amino acid metabolism, glutathione biosynthesis and resistance to oxidative stress37. The latter pathway was discovered based on its homology to the amino acid starvation control response in yeast (38 and Box 1).
Box 1: The integrated stress response (ISR).
In mammalia four eIF2α kinases, termed general control nonderepressible (GCN) 2, heme-regulated eIF2α kinase (HRI), protein kinase R (PKR) and PERK jointly constitute the integrated stress response, an ancient stress response that is highly conserved from yeast to mammals39, 40. It is known to modulate protein biosynthesis by integrating various types of stress signals, including ER stress, amino acid deprivation, infection with double stranded RNA viruses, and oxidative stress41. These diverse signals activate specific stress kinases, resulting in the phosphorylation of serine 51 of the alpha subunit of the eukaryotic initiation factor 2 (eIF2), which has GTPase activity. This modification changes the capacity of eIF2α to be recharged by the nucleotide exchange factor eIF2B, which subsequently leads to a reduction in the availability of active initiation complexes, and thus attenuates translation. In addition, the expression of proteins responsible for damage repair is increased, whereas translation of constitutively expressed mRNA is shut down by redirection of these mRNAs from polysomes to discrete cytoplasmic foci known as stress granules (SGs) for transient storage42. Defects in the ISR are associated with several diseases including those caused by viral infections, diabetes and Alzheimer’s disease39, 40.
Detection of stress and activation of the UPR
Facing the ER lumen with their sensor domains are PERK, ATF6 and IRE1 which can sense ER stress in different ways. Originally, it was proposed that ER stress was perceived as a drop in the amount of free BiP (binding immunoglobulin protein, also known as Grp78 or Hspa5), the most abundant ER chaperone43, 44. BiP binds all three sensors but detaches upon ER stress to assist the folding of nascent proteins43, 44. Consistent with this model, release of BiP would be sufficient to allow formation of high molecular mass complexes of activated PERK and IRE143. However, there were a few limitations to this model45, 46 and alternatively, it was suggested that unfolded proteins bind IRE1 directly. Crystallization of yeast IRE1 revealed the presence of a peptide-binding groove in the luminal domain of active oligomerized IRE1 that was very reminiscent of the one present in MHC-molecules47. Binding of unfolded proteins to the inactive conformation triggered a conformational change in the sensor domain, opening the peptide groove and inducing oligomerization16, 48. In this latter model, it is thought that the function of BiP is to keep PERK and IRE1 in a free, monomeric state (reviewed in16). The crystal structure of the PERK luminal domain has not been solved yet, but based on structural and functional homology with IRE1, PERK could also be activated by higher order oligomerization49. How ATF6 senses unfolded proteins remains as yet unknown. BiP dissociates from ATF6 upon ER stress44 by an active regulatory mechanism, and this leads to a conformational change from an oligomeric to monomeric form50.
The UPR as a part of normal cellular physiology
While most of the signaling cascades of the UPR have been unraveled in conditions of ER stress - defined as an excess of client load to folding capacity – most likely this does not represent the prime function of the UPR51. Especially in vertebrates with complex secretory functions, the UPR provides flexibility to ensure the multitude of ER functions across a wide range of physiological demands. If the UPR would be launched only after misfolded proteins accumulate in the ER, this would be a highly inefficient system, causing unnecessary impairment of the ER before homeostasis is restored. An anticipatory response would be more desirable, especially when high ER client load handling is part of normal physiology51. In some immune cells, activation of the UPR is part of the normal differentiation program of a cell. The differentiation of antigen triggered B cells into antibody-producing plasma cells requires the expansion of the ER membrane, a response previously assumed to be triggered by the accumulation of unfolded heavy chains52. However, more recent data showed that the activation of the UPR occurs before the onset of Ig chain synthesis53 and even in the (engineered) absence of Ig molecules altogether54. Reporter gene studies in vivo showed constitutive activation of the IRE1 pathway in specific cell types, like CD8α+ dendritic cells and developing B and T cells without any sign of activation of any of the other UPR cascades55, 56. Similarly, activation of Toll-like receptors (TLR) impinges on UPR signaling cascades57, 58. This most likely reflects an anticipatory response of the cell to prepare the ER to combat infection. It is at present still unclear how IRE1 or other UPR sensors are activated in these conditions, however perturbations in the composition of the ER lipid bilayer have been demonstrated to directly trigger IRE1 and PERK independently of their luminal sensor domain59. In addition, the flavonoid component quercetin activates IRE1 by binding to a pocket at the dimer interface of the RNase domain60. Thus, alternative ways to trigger ER stress sensors exist. These act independently of defects in protein folding homeostasis.
The UPR shows conserved functions in immune responses
As IRE1 is the only UPR sensor in yeast, and HAC1 its only known target, it was long assumed that the most ancient function of the UPR was a transcriptional response intended to increase ER folding capacity. However, further data challenged this concept. In most fungi, the IRE1-HAC1 branch is conserved (Fig. 2)61. However in S. pombe, no HAC1 gene could be found and activation of IRE1 mainly leads to RIDD. RIDD may thus be the ancestral function of IRE115. In pathogenic fungi, a fully competent UPR is linked to pathogen virulence, facilitating secretion of toxic compounds or mediating adaptation of the fungus to a host microenvironment that triggers a UPR62.
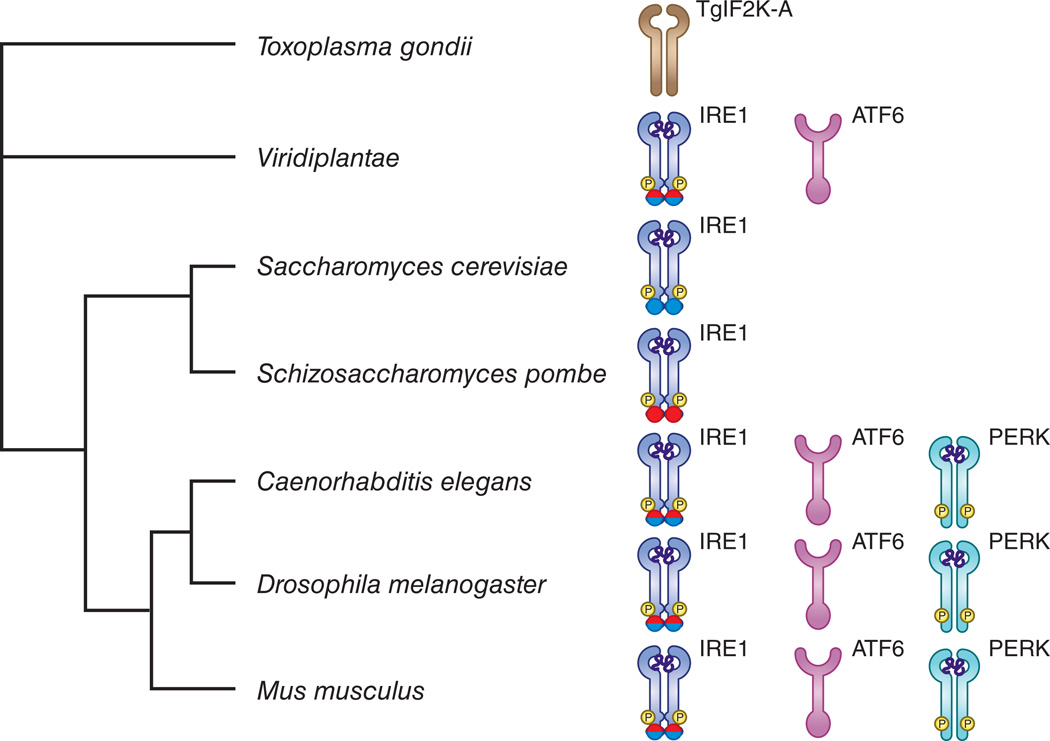
Figure 2. Evolution of the UPR shows conserved functions in immune responses.
Eukaryotic phylogenetic tree of the main species mentioned in the text. In S. pombe and S. cerevisiae only the IRE1 branch is present. In S. pombe the IRE-1 endonuclease is involved in RIDD (red color in endonuclease domain), in S. cerevisiae the IRE1 endonuclease is needed for splicing of the yeast XBP-1 homolog HAC-1 (blue color in endonuclease domain). In most fungi, a fully competent UPR is required for pathogen virulence. In protozoa, no recognizable orthologs of IRE1 and XBP1 can be found, however, some of them express an ER-based transmembrane kinase, called TgIF2K-A, that has the ability to phosphorylate eIF2α in response to ER stress, and exert some degree of translational control141. In plants, 2 branches of the UPR, ATF6 and IRE1, are represented. The plant UPR is also activated in response to pathogens and is needed for proper antibacterial defense. In C.elegans, the three branches of the UPR are found and mediate protection against overwhelming hyperinflammatory reactions. In mice and humans the three branches of the UPR are fully established and interact at different levels with inflammatory pathways.
In plants, the ATF6 and IRE1-XBP1 branch of the UPR appear to be conserved, while PERK is absent. In A. thaliana, the transcription factor bZIP60 is spliced by one of two IRE1 homologs at two stem-loop structures, which leads to the removal of a transmembrane domain and the nuclear localization of bZIP6063. Two ATF6 like factors, bZIP28 and bZIP17 are cleaved by S1P and S2P in response to ER stress and mediate the induction of BiP64. In addition, ER stress-induced downregulation of mRNAs has been observed65, and this RIDD-like mechanism was suggested to compensate for the lack of translational regulation by PERK. Intriguingly, in plants the UPR is not only activated in response to increased ER client protein load (e.g. during seed development), but also in response to abiotic stress, like heat shock or salt stress. In addition, IRE1 and bZIP60 is activated in response to plant pathogens and is required for antibacterial defense66, 67. Also in plants, the UPR response is anticipatory. During plant pathogen responses, foldases and chaperones are produced before defensive hydrolases are secreted68.
In Caenorhabditis elegans, the three branches of the UPR are conserved69. The IRE1-XBP-1 branch is specifically triggered by p38 homolog pmk in response to infection and is essential for larval survival in response to pathogens70. XBP-1 did not control the infection itself but rather was needed to prevent the detrimental effect of an overwhelming innate immune response on ER fitness. This suggests an evolutionarily-conserved role of the IRE1-XBP-1 arm to mediate protection against ER toxicity caused by inflammatory pathways71.
While the different branches of the UPR became fully established at the metazoan level only in the face of increasing complexity in the secretory functions of the ER, the integrated stress response is much older34, 61. Translational control as a proper response to nutrient deprivation was already established in yeast34, and most of the UPR sensors developed from more ancient metabolic regulators. As such, the PERK sensor is closely related to the GCN2 and GCN4 systems in unicellular eukaryotes and the regulated intramembrane proteolysis of ATF6-like factors is very similar to the system in which SREBP transcription factors control cholesterol metabolism27, 37.
Intriguingly, the integrated stress response and the UPR appear closely intertwined with host immune responses. Several microarray studies have shown a marked, though partial, overlap between genes induced by the UPR or ISR and genes induced by microbial infection or stimulation with TLR ligands72, 73. Compelling data on the activation of the ISR by bacterial72 or viral74 infection showed that metabolic responses controlling translation and autophagy most likely represent an ancient host defense response against invasive pathogens, that might predate the development of more dedicated pattern recognition receptors belonging to the NLR or TLR family72, 75.
The IRE-1-XBP-1 branch in differentiation of immune cells
In cells that have mainly a secretory function it is no surprise that genetic deletion of UPR sensors or their signaling intermediates also affects the cellular differentiation process. The analysis of PERK deficient mice revealed a crucial role for the UPR in exocrine pancreas acinar cells, endocrine pancreas insulin producing β-cells, chondrocytes and osteoclasts76, 77. XBP-1 plays a similar regulatory role in the development of exocrine pancreas acinar cells and intestinal Paneth cells78, 79. In the immune system, XBP-1 is essential in the terminal differentiation of B cells to plasma cells. It controls the expansion of the ER and its secretory function to enable the massive increase in immunoglobulin synthesis80,81. In B cells, the absence of XBP-1 led to hyperactivation of IRE-1, triggering RIDD and decay of secretory immunoglobulin µ mRNA, further curtailing IgM responses82. XBP-1 turned out to be downstream of Blimp-1 and IRF4, and was proposed to be the central hub in the physiological UPR of plasma cells11.
Further studies with RAG-2 blastocyst complementation systems showed that XBP-1 is also needed for the development and survival of other immune cell types, such as splenic conventional and plasmacytoid dendritic cells (cDCs and pDCs respectively)83. By making use of an IRE-1 activity reporter mouse (termed ERAI84), one study could confirm the constitutive but subset-specific activation of the IRE1 branch in CD8α+ cDCs55. Splenic macrophages and NK cells also activate the IRE1-XBP-branch at baseline, while naive T and B cells, monocytes and neutrophils do not show any basal IRE1 activity55. The physiological role of IRE1-XBP-1 in certain DC populations at baseline is still unclear. Spontaneous UPR could be linked to the capacity of pDCs to produce type I interferon83. In CD8α+ cDCs, it could be related to antigen cross-presentation, as well as to an adaptive strategy to combat infection55.
The UPR triggers inflammation
Defects in protein folding, either environmentally induced or caused by genetic defects in individual branches of the UPR, spontaneously induce an inflammatory phenotype. This has been described particularly in models of inflammatory bowel disease (IBD)71, metabolic disease85 or lung respiratory disease86. Loss of XBP-1 in intestinal epithelial or Paneth cells leads to enteritis, associated with strongly increased nuclear factor κB (NF-κB) activation. This is dependent on the hyperactivation of IRE-1 (and consequent RIDD), rather than on the loss of XBP-171. Interestingly, polymorphisms in both XBP1 and AGR2, a protein disulfide isomerase needed for mucin folding, have been found in Crohn’s disease and Ulcerative Colitis patients, suggesting that unresolved ER stress due to improper functioning of major UPR branches could indeed contribute to the inflammatory pathology typically observed in IBD87. This and many other studies, especially in diabetes85, 88, have led to the paradigm that dysregulated UPR signaling underlies chronic low-grade inflammation89.
The UPR intersects at various levels with inflammatory pathways, such as reactive oxygen species (ROS) production, and the activation of NF-κB, c-Jun N-terminal kinase (JNK), and IRF3 (Fig, 3 and Fig. 4)90. Activation of PERK and concomitant translational inhibition leads to a disequilibrium in the ratio of the short-lived IκB protein (inhibitor of NF-κB) to the longer-lived NF-κB protein, resulting in activation of NF-κB, independent of IκB phosphorylation91. On the other hand, IRE-1 directly triggers IκB kinase and as such IκB phosphorylation, in a TNF receptor associated factor (TRAF)-2 dependent manner92, 93, while ATF6 activates NF-κB via AKT phosphorylation94. IRE-1 also mediates phosphorylation of JNK in a TRAF2-ASK dependent pathway18, which was linked to insulin receptor substrate (IRS)-1 phosphorylation and the development of insulin resistance in type II diabetes95. ER stress and lipids also trigger the eIF2α-kinase PKR that coordinates activation of JNK and IRS-1 phosphorylation in a complex termed the ‘metaflammasome’85, 96. Also other innate immune pathways leading to IRF3 activation become activated in response to ER stress97.
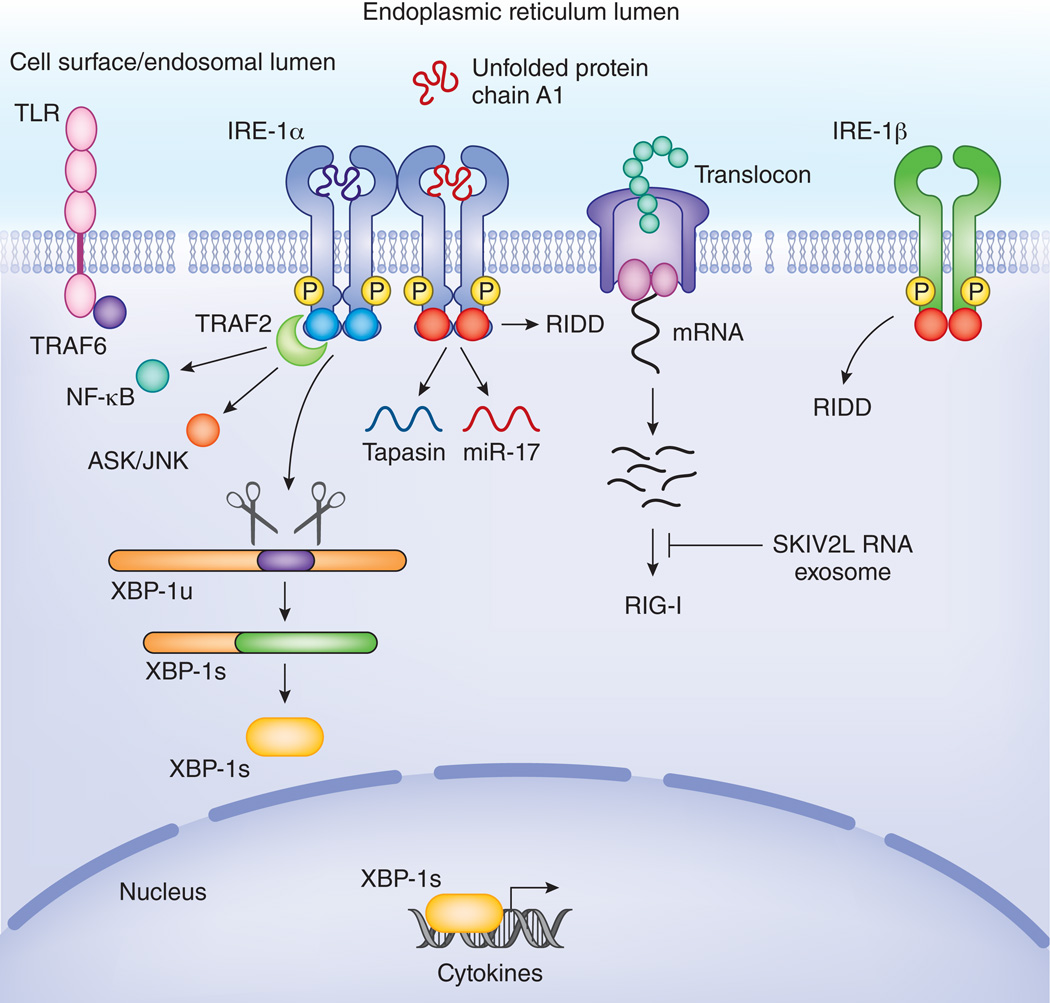
Figure 3. IRE1 intersects with inflammatory pathways.
TLRs stimulate IRE1 mediated XBP-1 splicing in a TRAF6 and NOX2 dependent manner. XBP-1s binds at the promoter of several cytokines and is needed for optimal cytokine expression. Phosphorylated IRE1 interacts via TRAF2 with the IKK complex and with ASK/JNK. In this way, IRE1 controls activation of NF-κB and AP-1, 2 major inflammatory transcription factors. IRE1 also serves as a host defense receptor, surveying the lumen of the ER for pathogens. In response to cholera toxin, IRE1 is activated and starts degrading mRNAs at the translocon complex. This generates several short RNA stretches that ligate RIG-I and trigger IFN-α via RIG-I. These RNA species can in turn be degraded by the SKIV2L exosome to prevent inappropriate inflammatory responses. RIDD also leads to degradation of specific mRNAs like that encoding tapasin as well as microRNAs such as miR-17. Degradation of tapasin and other components of the MHC-I loading machinery interfere with antigen presentation. Degradation of miR17, leads to degradation of TXNIP and subsequent stabilization of the NLRP3 inflammasome which causes enhanced IL-1β release. In addition to IRE-1α, a second isoform called IRE-1β is specifically expressed in epithelial cells lining mucosal interfaces, such as bronchi or the colon. Its RIDD function appears to predominate its capacity to splice XBP-1. This might endow IRE-1β with an enhanced capacity to exert immune surveillance and control the interface between the host and microbes present in the colon or airways.
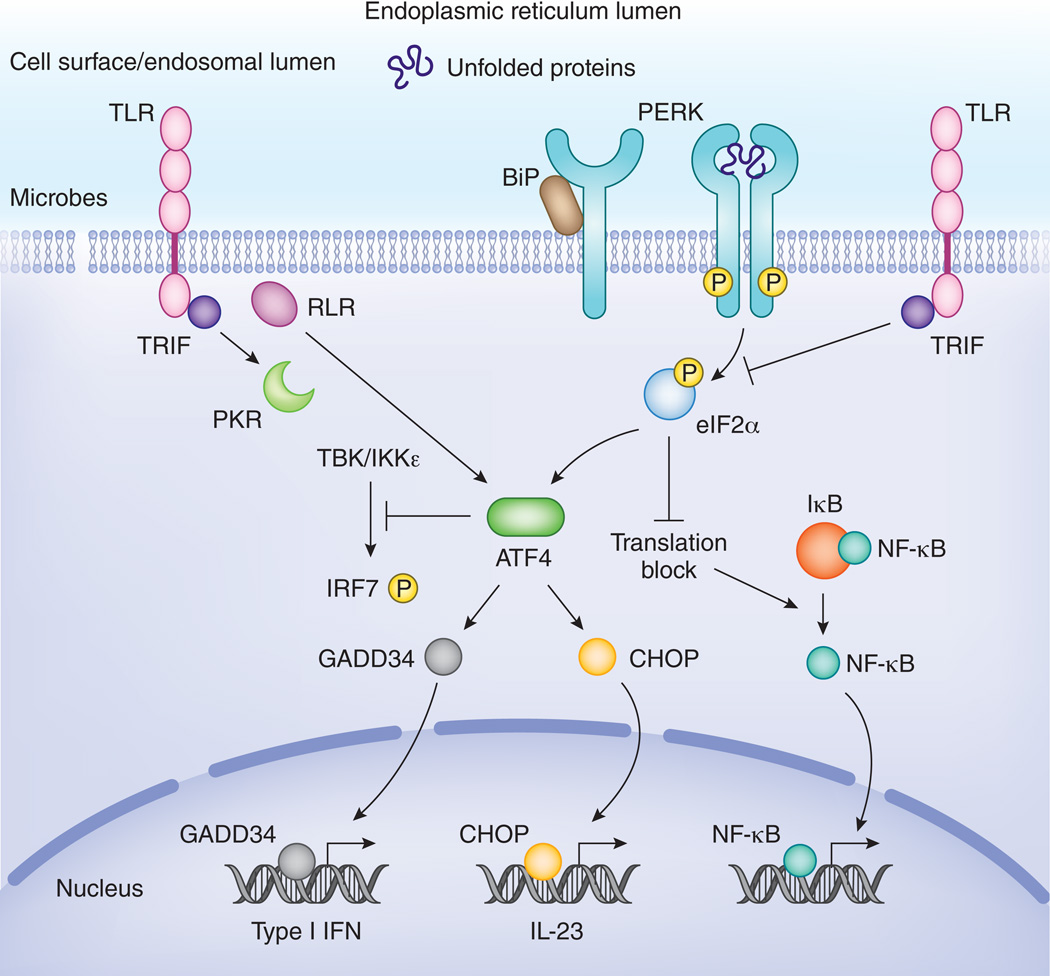
Figure 4. Pathways downstream of PERK are tightly controlled by inflammatory signals.
PERK-mediated translational inhibition leads to a shutdown of IκB de novo synthesis and as such leads to activation of NF-κB. ATF4 is involved in IFN and IL-23 cytokine expression via activation of GADD34 and CHOP respectively. On the other hand, it has also been reported that ATF4 interferes with TBK-IKKε mediated phosphorylation of IRF7 and IFN production in embryonic fibroblasts. TLRs tightly control the ATF4-CHOP branch and prevent induction of CHOP in macrophages, in a TRIF-dependent manner. In general, several microbial stress signals also lead to activation of ATF4, ATF3, CHOP or GADD34 in a PERK-independent, but TRIF, PKR or RLR-MAVS dependent manner. This microbial stress response hijacks components of the UPR, but has a different functional outcome.
There is limited evidence that the UPR by itself is sufficient to trigger production of inflammatory mediators such as interleukin-6 (IL-6), IL-8 or the proangiogenic factor VEGF. This appears highly cell type specific and can be mainly observed in cancer cells, epithelial or endothelial cells98, 99, 100. In DCs, the induction of IL-6 by tunicamycin was observed at the transcriptional but not at the protein level (Tavernier et al., unpublished data). Increased production of IL-1β upon UPR activation has been suggested to rely on IRE-1α mediated degradation of the micro RNA miR-17, causing the stabilization of thioredoxin interacting protein (TXNIP), and activation of the NLRP3 inflammasome101.
Several reports have shown that cytokine secretion by stimulation of the UPR requires a second signal57, 102. Concomitant activation of the UPR clearly amplifies the cytokine response induced by several TLR ligands57, 73, 103, 104, 105. According to the specific stimulus, this synergism depends on the IRE1-XBP-1s axis57, CHOP103 or the ATF4-GADD34-axis73. Synergism can be regulated at the transcriptional level, entailed by binding of XBP-1s106 or CHOP103 to particular cytokine promoters, but can also be regulated at the translational level73.
ER stress can also make cells refractory to inflammatory stimuli107, 108. For example it was proposed proposed that NF-κB activation resulting from ER stress occurs in a biphasic manner, with an early positive feedback loop and a later negative feedback loop109. Inhibition of NF-κB mediated signaling was shown to depend on TRAF2 downregulation90 and on induction of C/EBP-β by ER stress110, which acts as a transcriptional repressor of cytokine expression. This was confirmed in a recent study showing C/EBP-β dependent downregulation of IL-4 and IL-13 signaling upon ER stress111. Also other inflammatory pathways can be downregulated by UPR signaling components. ATF4 inhibits type I IFN expression, by interfering with TBK-IKKε-mediated phosphorylation of IRF7112.
Pathogens trigger the UPR
Both in plants and in C.elegans, infection is sufficient to trigger rigorous activation of UPR pathways66, 70. Also in mammals, several viruses, invasive bacteria and parasites elicit ER stress responses113, 114. Many pathogens interfere with the function of the ER as part of their infectious life cycle. Viruses like hepatitis C virus (HCV), poliovirus, human cytomegalovirus or herpes simplex virus strictly depend on the ER for assembly and budding of virion particles112. For successful replication of bacteria like Brucella or Legionella pneumophila and parasites like Toxoplasma inside the host cell, extensive interactions are required between the ER and the membranous compartments in which these pathogens reside114. Some bacterial toxins, like Shiga toxin, are retrogradely transported to the ER and as such interfere with ER homeostasis114. Unfolding of the A1 subunit of cholera toxin in the ER activates IRE1 and triggers the RIDD pathway, leading to the accumulation of small single stranded RNA products that launch a RIG-I dependent immune response115 (see Box 2).
Box 2: RIDD - an ancient IRE1 function?
The RNAse domain of D. melanogaster IRE-1α was found to have a more promiscuous function than just splicing of HAC1 or XBP16. Upon prolonged ER stress, IRE1 degrades diverse ER-localized mRNAs via RIDD. Though this function of IRE1 is now widely accepted and has been recognized in many different phyla, its exact physiological role and its regulation remain largely unclear128, 129. The free 5’ and 3’ ends of the RIDD-derived mRNAs become substrates for cellular exoribonucleases and as such are targeted for degradation23. It is still enigmatic what determines recognition by the IRE-1 endonuclease domain, but based on all validated substrates to-date a consensus sequence is emerging that shows similarity with the cleavage site in the stem loop structure of XBP-1 and that is highly conserved128. IRE-1 also degrades several miRNAs, and as such indirectly affects the expression of many hundreds of mRNAs130.
The physiological roles for RIDD have mainly been uncovered in genetic models of XBP-1 deficiency that lead in most - but not all - cells to strong activation of the RIDD pathway55, 131, 132. RIDD has a cytoprotective function in pancreatic beta cells in where it reduces the secretory load by degrading proinsulin131, or in liver cells by mediating protection from acetaminophen toxicity through the degradation mRNAs encoding cytochrome P450 enzymes132. In other systems, RIDD appears to play a cytotoxic role and it has been proposed that in conditions of unmitigated ER stress, RIDD triggers apoptosis17, 130, 133. In macrophages cholera toxin triggers the RIDD pathway upon unfolding of the A1 chain115. This leads to a rapid activation of the RIG-I pathway and a strong inflammatory cytokine response means IRE1α is the first pathogen receptor found surveying the lumen of the ER115. Further, the RNA helicase SKIV2L normally metabolizes intracellular RNA ligands generated upon activation of the UPR and as such limits activation of RIG-I134. In patients with genetic deficiencies in these RNA-metabolizing enzymes, a chronic inappropriate antiviral response would be triggered leading to gradual immune-mediated destruction of secretory cells with high constitutive UPR.
IRE-1β is typically expressed in epithelial cells lining mucosal interfaces, such as bronchi or the colon86. Its predominant RIDD activity might endow this molecule with an enhanced capacity to exert immune surveillance and control the interface between the host and many trillions of microbes present in the colon or airways115.
As some pathogens hijack the ER during their life cycle for assembly or exit, it is easy to understand how they activate the UPR. However, in other settings the mechanism is less obvious. Listeria monocytogenes has been found to induce ER expansion prior to host entry116, while HCV activates the UPR independent of viral replication. A direct interaction between HCV viral hydrophobic non-structural proteins and IRE-1 was proposed to trigger activation of the IRE1-XBP-1 arm117. Finally, stimulation of TLR2 and TLR4 by bacterial ligands in macrophages has been demonstrated to trigger specifically the IRE1-XBP-1 arm by a TRAF6 and NADPH oxidase 2 (NOX2) -dependent mechanism57.
Pathogen associated molecular patterns often activate a specific arm of the UPR, while actively suppressing the other arms118, 119, 120. TLR stimulation in macrophages induces XBP-1 splicing, but inhibits PERK or ATF6 activation57. Furthermore, LPS induces no canonical XBP-1s gene signature, but specifically triggers XBP-1s for optimal cytokine secretion57. This has also been observed in the case of HCV, CMV or West Nile virus infection117, 121, 122. TLR signaling suppresses the ATF4-CHOP branch by enhancing eIF2B guanine exchange activity and as such counteracting the inhibitory effects of phosphorylated eIF2α on ternary complex formation and protein translation58, 123. This TLR effect occurs via the adaptor molecule TRIF and allows for uninterrupted protein synthesis in response to pathogen infection in innate immune cells123. As a consequence, LPS pretreatment also prevented ER stress-induced CHOP expression and hence apoptosis, while GADD34 expression remained unaffected123. This was confirmed in other studies124, although the mechanism is likely to be more complex. In DCs for example, stimulation with lipopolysaccharide (LPS) does induce CHOP expression103, while stimulation with the TLR3 ligand poly(I:C) leads to upregulation of CHOP mRNA, but not protein73. The timing of LPS or poly(I:C) addition to DC cultures and UPR treatment determines whether CHOP is induced or not. Pretreatment with microbial stimuli appears to block subsequent UPR signals, while post-treatment with microbial stimuli leads to synergistic enhancement of the UPR response and CHOP expression, without causing cell death (Tavernier et al., unpublished data). This suggests that there are multiple mechanisms to modulate the UPR during immune responses. Induction of ATF4, ATF3, CHOP or GADD34 by microbial triggers can also occur in a PERK-independent manner, via TRIF, PKR or RIG-like receptor-MAVS pathways125. Furthermore, activation of these molecules downstream of the UPR or downstream of microbial stimulation yields a completely different functional outcome. Hence, pIC poly(I:C)-induced GADD34 is not sufficient to relieve the block on translational inhibition in immune cells, but rather plays a role in selective induction of IFN73. All these studies point towards an intimate link between pathogen detection, translation regulation and the UPR or ISR. It has been proposed that the term “microbial stress response” should be used to define these stress pathways. They are activated by pathogens, hijack some of the main components of the UPR for slightly adapted functions, without fully activating all arms of the UPR125.
Subversion of the UPR by viruses
There are several reasons why viruses would benefit from triggering the UPR. An increase in folding capacity and chaperones could sustain viral replication. Activation of lipid biosynthesis pathways through the UPR could help the formation of membrane associated replication complexes113. However, the UPR can also have adverse effects on viral propagation. Both the PERK dependent block in protein translation, the IRE-1-RIDD dependent degradation of glycoprotein-encoding mRNAs, the induction of IFN and the degradation of nascent viral proteins by ERAD pathways are likely to have a negative impact on viral replication113. Emerging evidence indicates that viruses selectively modulate specific branches of the UPR to maximally benefit from the UPR while circumventing the detrimental effects. Hepatitis C virus (HCV) triggers the UPR, which in turn activates the autophagic pathway that promotes viral replication126. The cytomegalovirus protein M50 specifically constrains IRE1-dependent ERAD pathways by binding to and degrading IRE1121. On the other hand, the IRE1 pathway is specifically activated by Japanese encephalitis virus (JEV) and influenza to support viral replication119, 127. In the case of JEV, the beneficial effect of IRE1 was found to depend on activation of the RIDD pathway127. RIDD led to cleavage of host transcripts, without any effect on the JEV RNA whereas inhibition of RIDD activity led to a reduction in JEV viral protein translation by an unknown mechanism127. In contrast, IRE1-dependent RIDD has been shown to degrade Respiratory syncytial virus (RSV) mRNAs and as such IRE1 activation blocks RSV replication118. How RIDD activity and target selection is regulated is far from clear (Box 2).
ER stress pathways and antigen presentation during vaccination
There is a generally unappreciated link between activation of the UPR and ISR pathways in DCs and antigen presentation to CD4 and CD8 T cells. XBP1 binds to a cyclic AMP-response element (CRE)-like sequence in the genes encoding MHC class II HlA-DPB and HlA- DRA135. Also H2-M2, a murine MHC class Ib gene with as yet unknown function, appears to be dependent on XBP-1 expression55. Several studies have shown that ER stress interferes with MHC-I surface expression, most likely linked with a defect in antigen supply due to an inhibition in protein translation136, 137. For example, activation of RIDD, but not loss of XBP-1s, interferes with the crosspresentation ability of CD8α DCs due to the degradation of several components in the crosspresentation machinery, notably tapasin55. Since CD8α DCs show high constitutive IRE1 endonuclease activity, it is possible that RIDD represents an alternative mechanism to prevent constitutive presentation of autoantigens and as such avoid autoimmunity. Phosphorylation of the PERK-eIF2a branch favors the translation of so-called cryptic antigens, initiated by CUG rather than AUG-codons138. As such, ER stress might change the peptide repertoire presented by MHC molecules, an area that certainly deserves further attention.
Finally, studies using systems biology approaches in humans have revealed that the live attenuated yellow fever vaccine 17D induced the expression of GCN2 in peripheral blood mononuclear cells within a few days after vaccination. This expression strongly correlates with the magnitude of the later CD8+ T cell response to the vaccine139. Subsequent mechanistic studies using GCN2 deficient mice demonstrated a critical role for virus-induced GCN2 activation in programming DCs to initiate autophagy and enhance antigen presentation to both CD4+ and CD8+ T cells74. These results reveal an unappreciated link between virus-induced ISR in DCs and the adaptive immune response. Furthermore, these results suggest that vaccine adjuvants that activate GCN2 in DCs may be efficient at inducing enhanced antigen presentation to T cells. Induction of the ISR or UPR might be predictive of good vaccine responses in humans. Consistent with this notion, the same vaccinology study showed that in human subjects vaccinated against influenza, the early expression of XBP-1 and other genes related to the UPR are robust biomarkers for the later occurrence of protective antibody titers139. In cancer, immunogenic cell death is favorable for anti-tumor responses and is accompanied by expression of the ER chaperone calreticulin on the cell surface of dying cells. The expression of calreticulin was shown to depend on PERK140.
Concluding remarks
The UPR is more than just an adaptive response to accumulation of unfolded proteins in the ER. In some immune cells, like plasma cells and DCs, as well as in barrier epithelial cells that are increasingly implicated in the regulation of the mucosal immune response, parts of the UPR and the ISR seem to be crucial for normal cellular differentiation and function. In the coming years, there will be a need to explore in greater detail how the UPR, or selective arms of this response, are activated as part of this normal physiology. It is possible that lineage-specific transcription factors drive the expression of certain UPR sensors, but this needs more study. Alternatively, some specialized cellular functions like crosspresentation require poorly studied cell biological processes, like the fusion of ER components with phagosomes. This might also lead to exploitation or triggering of certain UPR pathways like ERAD, or expansion of ER membranes or ER to Golgi transport. The degradation of RNA by IRE1 (RIDD) is emerging not only as a novel means of gene regulation in conditions of ER stress, but also could constitute an ancient pattern recognition pathway that senses ER resident pathogens and triggers immunity via the RIG-I pathway. It will be crucial to understand when exactly this pathway gets triggered and how pathogens subvert it.
Activation of innate immune receptors like TLRs or RIG-I receptors intersect with the UPR signaling pathways. We need to understand better if and how this represents a cellular adaptation to prepare the cell for secretion of cytokines, inflammatory mediators or other defense mechanisms. Given the fact that genome-wide studies of genetic risk have identified key UPR regulators to be associated with the risk of asthma, IBD and diabetes, we also need to understand in much greater detail how alterations in the UPR lead to chronic inflammatory disease, and whether these UPR related polymorphisms mainly affect inflammatory or structural cells. Ultimately, interfering with the UPR might constitute a novel way of promoting immunity or alternatively to circumvent chronic immune activation. Much more research is needed before we can exploit this new knowledge to the design of better vaccines or forms of immunotherapy.
Acknowledgements
B.N.L. is a recipient of an ERC Consolidator grant. B.N.L and S.J. are holders of several FWO program grants. B.N.L and S.J. are recipient of a UGhent MRP grant (Group-ID). Bali Pulendran acknowledges the generous support from the U.S. National Institutes of Health (grants R37 DK057665, R37 AI048638, U19 AI090023, U19 AI057266) and from the Bill & Melinda Gates Foundation. We would like to thank Fabio Martinon (Université de Lausanne, Switzerland), Pieter de Bleser, Paco Hulpiau and Liesbet Martens (Bio-IT Core, IRC, UGhent, Belgium) for helpful comments for the bioinformatics analysis.
References
- 1.Araki K, Nagata K. Protein folding and quality control in the ER. Cold Spring Harb Perspect Bio. 2012;4(8):a015438. doi: 10.1101/cshperspect.a015438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334(6059):1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma Y, Hendershot LM. The stressful road to antibody secretion. Nat Immunol. 2003;4(4):310–311. doi: 10.1038/ni0403-310. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–274. doi: 10.1038/16729. First paper of a series of studies unraveling the role of PERK in regulating protein translation in response to ER stress. [DOI] [PubMed] [Google Scholar]
- 5.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186(3):323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313(5783):104–107. doi: 10.1126/science.1129631. First study to demonstrate the role of IRE1 in a previously unanticipated function to control mRNA degradation. [DOI] [PubMed] [Google Scholar]
- 7.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101(3):249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 8.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87(3):391–404. doi: 10.1016/s0092-8674(00)81360-4. The Walter and the Mori group (ref 10 and others) almost simultaneously discovered the role of XBP-1 in the unfolded protein response. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer AL, Shapiro-Shelef M, Iwakoshi N, Lee A, Qian S, Zhao H, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 12.Shoulders MD, Ryno LM, Genereux JC, Moresco JJ, Tu PG, Wu C, et al. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 2013;3(4):1279–1292. doi: 10.1016/j.celrep.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167(1):35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4(2):265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 15.Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragon T, et al. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife. 2012;1:e00048. doi: 10.7554/eLife.00048. Paper demonstrating that the ancestral function of IRE1 was most likely RIDD and not a transcriptional response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Bi. 2012;28:251–277. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh R, Wang L, Wang ES, Perera BG, Igbaria A, Morita S, et al. Allosteric Inhibition of the IRE1alpha RNase Preserves Cell Viability and Function during Endoplasmic Reticulum Stress. Cell. 2014;158(3):534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 19.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 20.Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, et al. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J Clin Invest. 2001;107(5):585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martino MB, Jones L, Brighton B, Ehre C, Abdulah L, Davis CW, et al. The ER stress transducer IRE1beta is required for airway epithelial mucin production. Mucosal Immunol. 2013;6(3):639–654. doi: 10.1038/mi.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuru A, Fujimoto N, Takahashi S, Saito M, Nakamura D, Iwano M, et al. Negative feedback by IRE1beta optimizes mucin production in goblet cells. Proc Natl Acad Sci U S A. 2013;110(8):2864–2869. doi: 10.1073/pnas.1212484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, et al. IRE1beta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7(5):445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwawaki T, Hosoda A, Okuda T, Kamigori Y, Nomura-Furuwatari C, Kimata Y, et al. Translational control by the ER transmembrane kinase/ribonuclease IRE1 under ER stress. Nat Cell Biol. 2001;3(2):158–164. doi: 10.1038/35055065. [DOI] [PubMed] [Google Scholar]
- 25.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. First paper of a series of studies that uncovered together the role of ATF6 in the UPR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273(50):33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 27.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 28.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33(1):75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 29.Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, et al. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122(Pt 10):1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo S, Saito A, Asada R, Kanemoto S, Imaizumi K. Physiological unfolded protein response regulated by OASIS family members, transmembrane bZIP transcription factors. IUBMB Life. 2011;63(4):233–239. doi: 10.1002/iub.433. [DOI] [PubMed] [Google Scholar]
- 31.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124(3):587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 32.Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18(12):7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 34.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor Perspect Bio. 2012;4(10) doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 36.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. Embo J. 2003;22(5):1180–1187. doi: 10.1093/emboj/cdg112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 38.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 39.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. 2013;70(19):3493–3511. doi: 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34(Pt 1):7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 41.Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutrition. 2012;3(3):307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19(10):R397–R398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiPand ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 44.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 45.Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8(7):e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol. 2004;167(3):445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102(52):18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333(6051):1891–1894. doi: 10.1126/science.1209126. Seminal paper presenting an alternative model for IRE1 activation through direct binding of unfolded proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275(32):24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 50.Nadanaka S, Okada T, Yoshida H, Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27(3):1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189(5):783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4(4):321–329. doi: 10.1038/ni907. One of the earliest papers showing an essential role for XBP-1 in immune cells. [DOI] [PubMed] [Google Scholar]
- 53.van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18(2):243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 54.Hu CC, Dougan SK, McGehee AM, Love JC, Ploegh HL. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. Embo J. 2009;28(11):1624–1636. doi: 10.1038/emboj.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osorio F, Tavernier SJ, Hoffmann E, Saeys Y, Martens L, Vetters J, et al. The unfolded-protein-response sensor IRE-1alpha regulates the function of CD8alpha dendritic cells. Nat Immunol. 2014;15(3):248–257. doi: 10.1038/ni.2808. The first paper demonstrating a role for the IRE1-XBP-1 branch in MHC-I antigen presentation in dendritic cells via the RIDD pathway. [DOI] [PubMed] [Google Scholar]
- 56.Brunsing R, Omori SA, Weber F, Bicknell A, Friend L, Rickert R, et al. B- and T-cell development both involve activity of the unfolded protein response pathway. J Biol Chem. 2008;283(26):17954–17961. doi: 10.1074/jbc.M801395200. [DOI] [PubMed] [Google Scholar]
- 57.Martinon F, Chen X, Lee A-H, Glimcher LH. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat Immunol. 2010;11(5):411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woo CW, Kutzler L, Kimball SR, Tabas I. Toll-like receptor activation suppresses ER stress factor CHOP and translation inhibition through activation of eIF2B. Nat Cell Biol. 2012;14(2):192–200. doi: 10.1038/ncb2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A. 2013;110(12):4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiseman RL, Zhang Y, Lee KP, Harding HP, Haynes CM, Price J, et al. Flavonol activation defines an unanticipated ligand-binding site in the kinase-RNase domain of IRE1. Mol Cell. 2010;38(2):291–304. doi: 10.1016/j.molcel.2010.04.001. First study demonstrating activation of IRE1 not via its luminal but via its cytoplasmic domain, suggesting the existence of alternative ways to trigger UPR sensors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hollien J. Evolution of the unfolded protein response. BBA. 2013;1833(11):2458–2463. doi: 10.1016/j.bbamcr.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 62.Askew DS. Endoplasmic reticulum stress and fungal pathogenesis converge. Virulence. 2014;5(2):331–333. doi: 10.4161/viru.28051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagashima Y, Mishiba K, Suzuki E, Shimada Y, Iwata Y, Koizumi N. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep. 2011;1:29. doi: 10.1038/srep00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell. 2007;19(12):4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez IM, Chrispeels MJ. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell. 2003;15(2):561–576. doi: 10.1105/tpc.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, Jordan MR, et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS One. 2012;7(2):e31944. doi: 10.1371/journal.pone.0031944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tateda C, Ozaki R, Onodera Y, Takahashi Y, Yamaguchi K, Berberich T, et al. NtbZIP60, an endoplasmic reticulum-localized transcription factor, plays a role in the defense response against bacterial pathogens in Nicotiana tabacum. J Plant Res. 2008;121(6):603–611. doi: 10.1007/s10265-008-0185-5. [DOI] [PubMed] [Google Scholar]
- 68.Jelitto-Van Dooren EP, Vidal S, Denecke J. Anticipating endoplasmic reticulum stress. A novel early response before pathogenesis-related gene induction. Plant Cell. 1999;11(10):1935–1944. doi: 10.1105/tpc.11.10.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107(7):893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 70.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463(7284):1092–1095. doi: 10.1038/nature08762. Elegant study demonstrating that in C. elegans XBP-1 tempers an otherwise devastating immune response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503(7475):272–276. doi: 10.1038/nature12599. One of several papers from unraveling the role of the IRE1-XBP-1 branch in mucosal immunology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tattoli I, Sorbara MT, Vuckovic D, Ling A, Soares F, Carneiro LA, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11(6):563–575. doi: 10.1016/j.chom.2012.04.012. Very elegant study demonstrating how the integrated stress response and innate host defense responses are very closely linked. [DOI] [PubMed] [Google Scholar]
- 73.Clavarino G, Claudio N, Couderc T, Dalet A, Judith D, Camosseto V, et al. Induction of GADD34 is necessary for dsRNA-dependent interferon-beta production and participates in the control of Chikungunya virus infection. PLoS Path. 2012;8(5):e1002708. doi: 10.1371/journal.ppat.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343(6168):313–317. doi: 10.1126/science.1246829. The first study to unravel the role of GCN2 in enhancing autophagy and antigen presentation in dendritic cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lemaitre B, Girardin SE. Translation inhibition and metabolic stress pathways in the host response to bacterial pathogens. Nat Rev Microbiol. 2013;11(6):365–369. doi: 10.1038/nrmicro3029. [DOI] [PubMed] [Google Scholar]
- 76.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7(6):1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 77.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, et al. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22(11):3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaser A, Lee A-H, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 Links ER Stress to Intestinal Inflammation and Confers Genetic Risk for Human Inflammatory Bowel Disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. Embo J. 2005;24(24):4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412(6844):300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 81.Todd DJ, McHeyzer-Williams LJ, Kowal C, Lee A-H, Volpe BT, Diamond B, et al. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J Exp Med. 2009;206(10):2151–2159. doi: 10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benhamron S, Hadar R, Iwawaky T, So JS, Lee AH, Tirosh B. Regulated IRE1-dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur J Immunol. 2014;44(3):867–876. doi: 10.1002/eji.201343953. [DOI] [PubMed] [Google Scholar]
- 83.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J Exp Med. 2007;204(10):2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10(1):98–102. doi: 10.1038/nm970. Development of a reporter gene tool that appeared extremely useful to monitor IRE1 activation in physiological conditions in vivo. [DOI] [PubMed] [Google Scholar]
- 85.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140(6):900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Osorio F, Lambrecht B, Janssens S. The UPR and lung disease. Semin Immunopathol. 2013;35(3):293–306. doi: 10.1007/s00281-013-0368-6. [DOI] [PubMed] [Google Scholar]
- 87.Adolph TE, Niederreiter L, Blumberg RS, Kaser A. Endoplasmic reticulum stress and inflammation. Dig Dis. 2012;30(4):341–346. doi: 10.1159/000338121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engin F, Yermalovich A, Nguyen T, Hummasti S, Fu W, Eizirik DL, et al. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5(211):211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hasnain SZ, Lourie R, Das I, Chen AC-H, McGuckin MA. The interplay between endoplasmic reticulum stress and inflammation. Immunol Cell Biol. 2012;90(3):260–270. doi: 10.1038/icb.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jiang HY, Wek SA, McGrath BC, Scheuner D, Kaufman RJ, Cavener DR, et al. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23(16):5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26(8):3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaneko M, Niinuma Y, Nomura Y. Activation signal of nuclear factor-kappa B in response to endoplasmic reticulum stress is transduced via IRE1 and tumor necrosis factor receptor-associated factor 2. Biol Pharm Bull. 2003;26(7):931–935. doi: 10.1248/bpb.26.931. [DOI] [PubMed] [Google Scholar]
- 94.Yamazaki H, Hiramatsu N, Hayakawa K, Tagawa Y, Okamura M, Ogata R, et al. Activation of the Akt-NF-kappaB pathway by subtilase cytotoxin through the ATF6 branch of the unfolded protein response. J Immunol. 2009;183(2):1480–1487. doi: 10.4049/jimmunol.0900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306(5695):457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 96.Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, et al. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140(3):338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petrasek J, Iracheta-Vellve A, Csak T, Satishchandran A, Kodys K, Kurt- Jones EA, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc Natl Acad Sci USA. 2013;110(41):16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gora S, Maouche S, Atout R, Wanherdrick K, Lambeau G, Cambien F, et al. Phospholipolyzed LDL induces an inflammatory response in endothelial cells through endoplasmic reticulum stress signaling. Faseb J. 2010;24(9):3284–3297. doi: 10.1096/fj.09-146852. [DOI] [PubMed] [Google Scholar]
- 99.Maguire JA, Mulugeta S, Beers MF. Endoplasmic reticulum stress induced by surfactant protein C BRICHOS mutants promotes proinflammatory signaling by epithelial cells. Am J Resp Cell Mol Biol. 2011;44(3):404–414. doi: 10.1165/rcmb.2009-0382OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marjon PL, Bobrovnikova-Marjon EV, Abcouwer SF. Expression of the pro-angiogenic factors vascular endothelial growth factor and interleukin-8/CXCL8 by human breast carcinomas is responsive to nutrient deprivation and endoplasmic reticulum stress. Mol Cancer. 2004;3:4. doi: 10.1186/1476-4598-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, Igbaria A, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16(2):250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu F, Yu X, Wang H, Zuo D, Guo C, Yi H, et al. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. Eur J Immunol. 2011;41(4):1086–1097. doi: 10.1002/eji.201040831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goodall JC, Wu C, Zhang Y, McNeill L, Ellis L, Saudek V, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci USA. 2010;107(41):17698–17703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martino MEB, Olsen JC, Fulcher NB, Wolfgang MC, O'Neal WK. Ribeiro CMP. Airway epithelial inflammation-induced endoplasmic reticulum Ca2+ store expansion is mediated by X-box binding protein-1. J Biol Chem. 2009;284(22):14904–14913. doi: 10.1074/jbc.M809180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smith JA, Turner MJ, DeLay ML, Klenk EI, Sowders DP, Colbert RA. Endoplasmic reticulum stress and the unfolded protein response are linked to synergistic IFN-beta induction via X-box binding protein 1. Eur J Immunol. 2008;38(5):1194–1203. doi: 10.1002/eji.200737882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dalod M, Pierre P. Integration of ER stress and viral nucleotide sensing in DCs: mounting a response commensurate to the threat? Eur J Immunol. 2011;41(4):898–901. doi: 10.1002/eji.201141497. [DOI] [PubMed] [Google Scholar]
- 107.Hayakawa K, Hiramatsu N, Okamura M, Yamazaki H, Nakajima S, Yao J, et al. Acquisition of anergy to proinflammatory cytokines in nonimmune cells through endoplasmic reticulum stress response: a mechanism for subsidence of inflammation. J Immunol. 2009;182(2):1182–1191. doi: 10.4049/jimmunol.182.2.1182. [DOI] [PubMed] [Google Scholar]
- 108.Li J, Wang JJ, Zhang SX. Preconditioning with endoplasmic reticulum stress mitigates retinal endothelial inflammation via activation of X-box binding protein 1. J Biol Chem. 2011;286(6):4912–4921. doi: 10.1074/jbc.M110.199729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitamura M. Control of NF-kappaB and inflammation by the unfolded protein response. Int Rev Immunol. 2011;30(1):4–15. doi: 10.3109/08830185.2010.522281. [DOI] [PubMed] [Google Scholar]
- 110.Hayakawa K, Nakajima S, Hiramatsu N, Okamura M, Huang T, Saito Y, et al. ER stress depresses NF-kappaB activation in mesangial cells through preferential induction of C/EBP beta. J Am Soc Nephrol. 2010;21(1):73–81. doi: 10.1681/ASN.2009040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arensdorf AM, Rutkowski DT. Endoplasmic reticulum stress impairs IL-4/IL-13 signaling through C/EBPbeta-mediated transcriptional suppression. J Cell Sci. 2013;126(Pt 17):4026–4036. doi: 10.1242/jcs.130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liang Q, Deng H, Sun CW, Townes TM, Zhu F. Negative regulation of IRF7 activation by activating transcription factor 4 suggests a cross-regulation between the IFN responses and the cellular integrated stress responses. J Immunol. 2011;186(2):1001–1010. doi: 10.4049/jimmunol.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.He B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006;13(3):393–403. doi: 10.1038/sj.cdd.4401833. [DOI] [PubMed] [Google Scholar]
- 114.Roy CR, Salcedo SP, Gorvel JP. Pathogen-endoplasmic-reticulum interactions: in through the out door. Nat Rev Immunol. 2006;6(2):136–147. doi: 10.1038/nri1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cho JA, Lee AH, Platzer B, Cross BC, Gardner BM, De Luca H, et al. The unfolded protein response element IRE1alpha senses bacterial proteins invading the ER to activate RIG-I and innate immune signaling. Cell Host Microbe. 2013;13(5):558–569. doi: 10.1016/j.chom.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 116.Pillich H, Loose M, Zimmer KP, Chakraborty T. Activation of the unfolded protein response by Listeria monocytogenes. Cell Microbiol. 2012;14(6):949–964. doi: 10.1111/j.1462-5822.2012.01769.x. [DOI] [PubMed] [Google Scholar]
- 117.Ambrose RL, Mackenzie JM. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J Virol. 2011;85(6):2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hassan I, Gaines KS, Hottel WJ, Wishy RM, Miller SE, Powers LS, et al. Inositol-requiring enzyme 1 inhibits respiratory syncytial virus replication. J Biol Chem. 2014;289(11):7537–7546. doi: 10.1074/jbc.M113.510594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, et al. Influenza A viral replication is blocked by inhibition of the inositolrequiring enzyme 1 (IRE1) stress pathway. J Biol Chem. 2012;287(7):4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tardif KD, Mori K, Kaufman RJ, Siddiqui A. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem. 2004;279(17):17158–17164. doi: 10.1074/jbc.M312144200. [DOI] [PubMed] [Google Scholar]
- 121.Stahl S, Burkhart JM, Hinte F, Tirosh B, Mohr H, Zahedi RP, et al. Cytomegalovirus downregulates IRE1 to repress the unfolded protein response. PLoS Pathog. 2013;9(8):e1003544. doi: 10.1371/journal.ppat.1003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng Y, Gao B, Ye L, Kong L, Jing W, Yang X, et al. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol. 2005;43(6):529–536. [PubMed] [Google Scholar]
- 123.Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA, et al. Adaptive suppression of the ATF4-CHOP branch of the unfolded protein response by toll-like receptor signalling. Nat Cell Biol. 2009;11(12):1473–1480. doi: 10.1038/ncb1996. One of the first papers to demonstrate that TLR signaling modifies the UPR response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakayama Y, Endo M, Tsukano H, Mori M, Oike Y, Gotoh T. Molecular mechanisms of the LPS-induced non-apoptotic ER stress-CHOP pathway. J Biochem. 2010;147(4):471–483. doi: 10.1093/jb/mvp189. [DOI] [PubMed] [Google Scholar]
- 125.Claudio N, Dalet A, Gatti E, Pierre P. Mapping the crossroads of immune activation and cellular stress response pathways. Embo J. 2013;32(9):1214–1224. doi: 10.1038/emboj.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121(1):37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bhattacharyya S, Sen U, Vrati S. Regulated IRE1-dependent decay pathway is activated during Japanese encephalitis virus-induced unfolded protein response and benefits viral replication. J Gen Virol. 2014;95(Pt 1):71–79. doi: 10.1099/vir.0.057265-0. [DOI] [PubMed] [Google Scholar]
- 128.Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39(5):245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 129.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 130.Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338(6108):818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lee A-H, Heidtman K, Hotamisligil G, Glimcher L. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci USA. 2011;108(21):8885–8890. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hur K, So J, Ruda V, Frank-Kamenetsky M, Fitzgerald K, Koteliansky V, et al. IRE1α activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209(2):307–318. doi: 10.1084/jem.20111298. Ref. 132 and 133 were the first to uncover hyperactivation of IRE1alpha and hence RIDD activity in the XBP-1 KO, explaining some of the observed phenotypes in the XBP-1 KO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138(3):562–575. doi: 10.1016/j.cell.2009.07.017. Study showing that with artificially designed IRE1 constructs it is possible to regulate RIDD and XBP-1 splicing activity independently. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, et al. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol. 2014 doi: 10.1038/ni.2948. Much like the exonuclease Trex1 metabolizing self DNA to avoid autoimmunity, the RNA exosome neutralizes RNA fragments generated by the RIDD pathway that trigger RIG-I receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liou HC, Boothby MR, Finn PW, Davidon R, Nabavi N, Zeleznik-Le NJ, et al. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science. 1990;247(4950):1581–1584. doi: 10.1126/science.2321018. [DOI] [PubMed] [Google Scholar]
- 136.Qian SB, Reits E, Neefjes J, Deslich JM, Bennink JR, Yewdell JW. Tight linkage between translation and MHC class I peptide ligand generation implies specialized antigen processing for defective ribosomal products. J Immunol. 2006;177(1):227–233. doi: 10.4049/jimmunol.177.1.227. [DOI] [PubMed] [Google Scholar]
- 137.Granados DP, Tanguay PL, Hardy MP, Caron E, de Verteuil D, Meloche S, et al. ER stress affects processing of MHC class I-associated peptides. BMC Immunol. 2009;10:10. doi: 10.1186/1471-2172-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2(11):e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–795. doi: 10.1038/ni.2067. Large system biology approach to uncover molecular signatures upon vaccination that correlated with and could be used to predict later antibody responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12(12):860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- 141.Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, et al. Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem. 2008;283(24):16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]