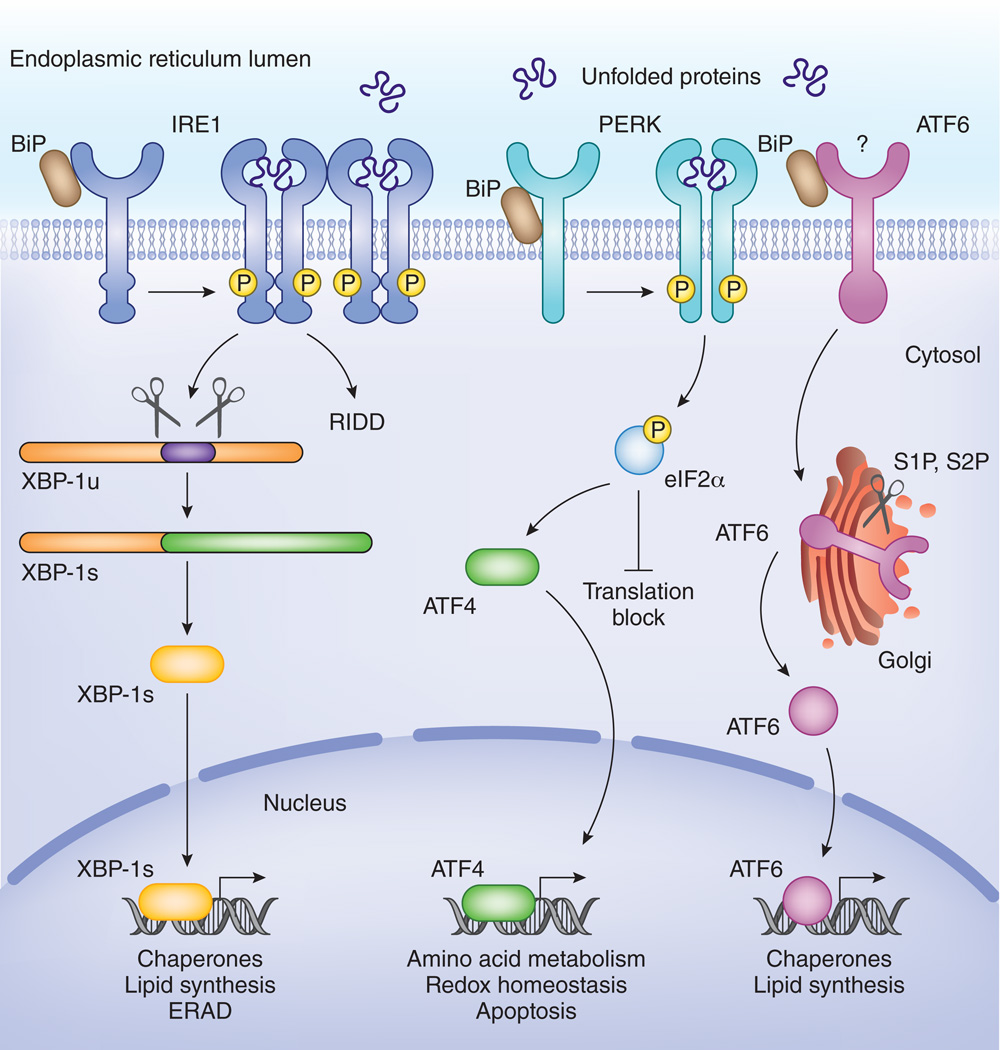
Figure 1. Three different sensors jointly coordinate the UPR in mammals.
IRE1 and PERK are activated by oligomerization and transphosphorylation upon binding of unfolded proteins and release of the chaperone BiP. The endonuclease domain of IRE1 cleaves XBP-1 mRNA in an unconventional splicing reaction to generate XBP-1s mRNA, and the resulting product encodes a transcription factor of the bZIP family. In addition the endonuclease activity of IRE-1 also contributes to Regulated IRE-1 dependent decay (RIDD), the degradation of mRNAs that are recruited at ribosomes at the ER. PERK is a transmembrane kinase that phosphorylates eukaryotic initiation factor 2 alpha (eIF2α). This leads to a transient inhibition of protein translation. Some mRNAs that encode small upstream open reading frames (uORFs) in their 5’ UTR escape the translation stop, the most prominent being the transcription factor ATF4. ATF4 in turn triggers expression of CHOP, GADD34 and additional factors important for amino acid metabolism and redox control. ATF6 is activated upon release of BiP and translocated to the Golgi where it undergoes sequential cleavage and removal of its luminal domain. The remaining transactivation domain of ATF6 moves to the nucleus and coordinates expression of genes involved in chaperone pathways or lipid biosynthesis.