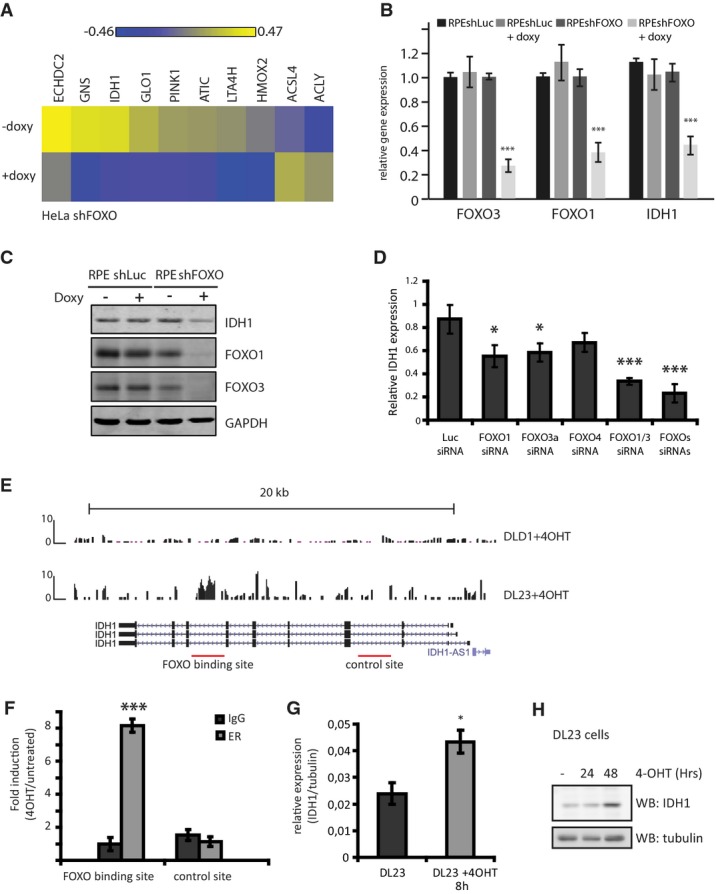
Heat map of selected metabolic genes showing differential expression in FOXO-depleted HeLashFOXO cells. HeLashFOXO cells were grown in the presence or absence of doxycycline for 72 h before RNA was collected and used for gene expression analysis. Results are represented as LIMMAS, comparing each sample against a common reference.
Relative FOXO3, FOXO1, and IDH1 mRNA levels in RPEshLuc and RPEshFOXO depleted or not of FOXOs. RPEshFOXO cells were cultured in the presence or absence of doxycycline for 72 h before sample collection (n = 3).
IDH1, FOXO1, and FOXO3 protein levels in RPEshLuc and RPEshFOXO depleted or not of FOXOs. Cells were grown in the presence or absence of doxycycline for 72 h before sample collection. Results are representative of at least three independent experiments.
Relative mRNA levels of IDH1 in RPE cells transfected with the indicated siRNAs. RNA samples were collected 72 h post-siRNA transfection. IDH1 expression was normalized by tubulin expression (n = 3).
ChIP-sequencing analysis identified a FOXO3 binding site in the IDH1 genomic region in DL23 cells.
ChIP–qPCR in DL23 cells 4 h after FOXO3 activation. The IDH1 (+) and the IDH1 (−) introns correspond to the FOXO binding site and the negative site in IDH1 (n = 3).
Relative IDH1 mRNA levels in DL23 cells. DL23 cells were induced with 4-OHT for 4 h before sample collection (n = 3).
IDH1 protein levels in DL23 cells grown in the presence of 4-OHT for different time points before sample collection. Result is representative of three independent experiments.
Data information: Unless stated otherwise, error bars represent standard deviation. Statistical analysis was performed with
-test. ***
< 0.05.