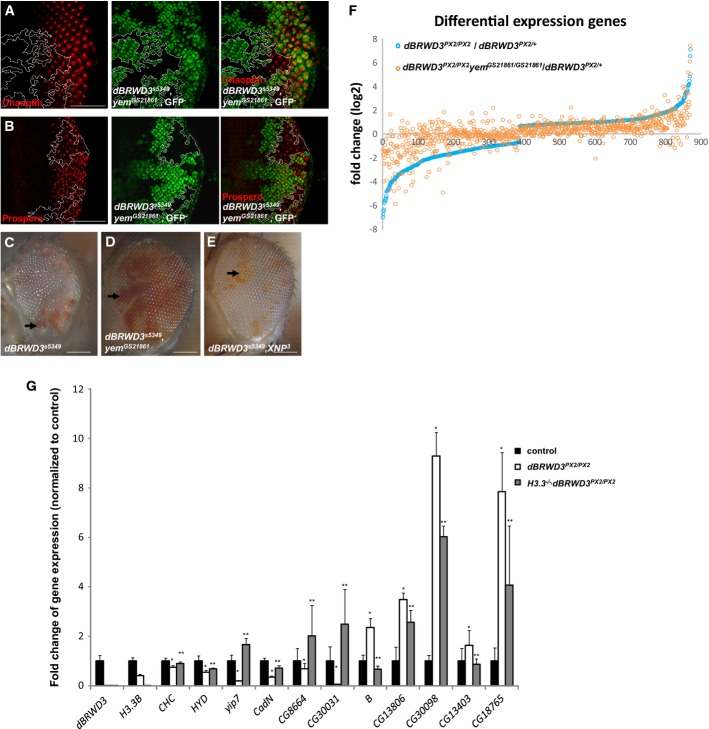
A, B Expression of neuronal genes, Chaoptin (A), and Prospero (B) (red) in dBRWD3s5349, yemGS21861 mutant photoreceptors marked by the absence of GFP. Scale bars indicate 50 μm.
C-E Suppression of the cell lethality in adult dBRWD3s5349 (null) clones by yemGS21861. Adult homozygous dBRWD3s5349 (C), dBRWD3s5349, yemGS21861 (D), and XNP3, dBRWD3s5349 (E) mutant photoreceptors were generated by ey-flp and marked by white gene expression (red regions, arrows), whereas wild-type twin clones were marked by the absence of white (white regions). Scale bars indicate 100 μm.
F Suppression of aberrant gene transcription in dBRWD3 mutant brains by yem mutation. A scatter plot of the fold changes of the differentially expressed genes in dBRWD3PX2/PX2 versus dBRWD3PX2/+ brains shown in an increasing order (blue circles). The corresponding fold changes in dBRWD3PX2/PX2, yemGS21861/GS21861 versus dBRWD3PX2/+ brains were shown as orange circles.
G Expression of the differentially expressed genes in dBRWD3PX2/PX2 and dBRWD3
PX2/PX2, H3.3B−/− brains. The expression of B, yip7, CadN, CG8664, CG13403, CG30031, CG13806, CG30098, CHC, HYD, and CG18765 in Canton S, dBRWD3PX2/PX2 and dBRWD3
PX2/PX2, H3.3B−/− brains was analyzed by RT–PCR and presented as mean fold changes from 4 experiments relative to Canton S ± 1 SD. *Indicates P < 0.05 versus Canton S and **indicates P < 0.05 versus dBRWD3PX2/PX2 by Student's t-test.