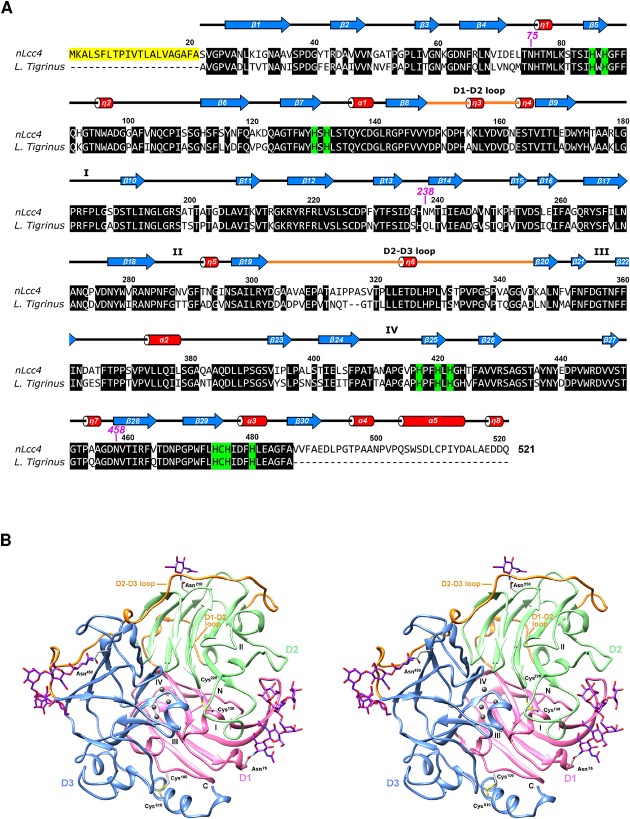
Fig 5. Structure-assisted sequence alignment and stereo views of the structure of nLcc4.
(A) Protein sequence alignment of Lentinus sp. Lcc4 (PDB #3X1B) and Lentinus tigrinus (PDB #2QT6). The signal peptide of Lentinus sp. Lcc4 is highlighted in yellow. Residues in α-helices, β -strands, and loops are shown as red cylinders, blue arrows, and lines, respectively. The 310-helices are labeled as η1 to η8. The histidine residues involved in the copper coordination are highlighted in green. The glycosylated Asn residues are marked as magenta numbers. The four substrate binding pocket loops (SBPLs I-IV) and the domain connection loops (D1-D2 and D2-D3 loops) are also indicated. (B) Ribbon diagram of laccase crystal structure, domain 1 (D1), domain 2 (D2) and domain 3 (D3) of laccase are colored pink, green and blue, respectively. Two disulfide bonds (Cys106-Cys510 and Cys138-Cys226) and three N-glycosylated sites (Asn75, Asn238, and Asn458) are labeled. The licorice representation shows: copper ions in spheres with dark grey and oligosaccharides in sticks with purple carbons. The predicted substrate binding pocket loops (SBPLs I-IV) are also indicated.