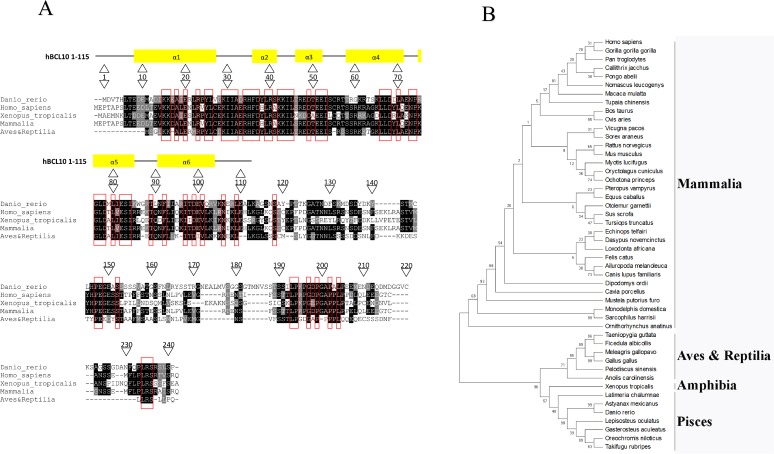
Fig 1. Alignment and phylogenetic tree of zBcl10.
A) Alignment of zBcl10 and human BCL10 with the consensus sequence generated by aligning the BCL10 sequences of fish, birds and reptiles, and mammals. The Xenopus tropicalis sequence is the only amphibian BCL10 sequence available. The alignment was done using ClustalW. Printout from multiple-aligned sequences and consensus sequences calculation were done with BOXSHADE. The black background designates identical amino acids, the gray background conservative substitutions. The red rectangles indicate amino acids conserved in all sequences examined. At the top of the alignment, the six alpha helix regions of the CARD are shown. The sequences used for alignment and generation of the consensus sequence are available in Supplementary Material. B) Phylogenetic tree analysis of BCL10 proteins. Phylogenetic analyses with bootstrapping (100 replicates) were obtained by the Neighbor-joining method using complete deletion and the p-distance amino acid model in MEGA [28]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) are shown next to the branches. The sequences used for phylogenetic tree generation are available in Supporting Information (S1 Table).