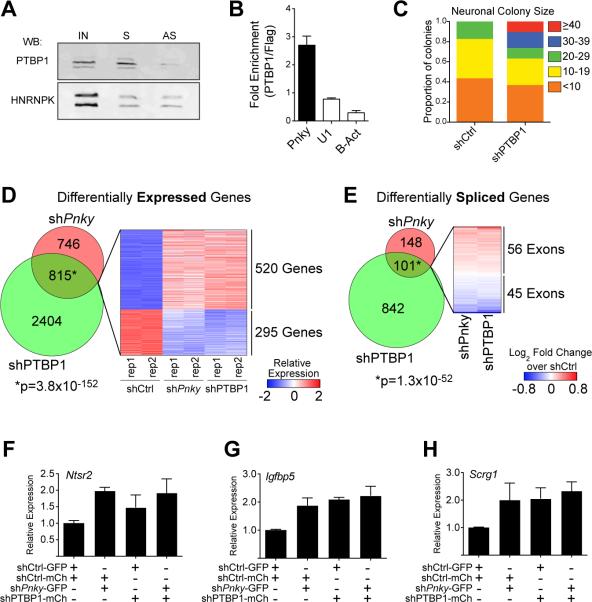
Figure 4. Pinky interacts with PTBP1 and regulates transcript expression and differential splicing.
A) Immunoblot for PTBP1 or HNRNPK following RNA-pulldown with biotin-labeled sense (S) or anti-sense (AS) Pnky RNA incubated with V-SVZ NSC nuclear extract. IN = input V-SVZ nuclear extract. B) RT-qPCR detection for indicated RNA recovered by PTBP1-specific antibody normalized to control FLAG antibody. Error bars are propagated S.D. from technical triplicates. C) Quantification of number of TUJ1+ neuroblasts found in individual neurogenic colonies with Ptbp1-knockdown or control. D) Venn Diagram demonstrating the overlap between genes differentially expressed (FDR < 0.05) upon Ptbp1 or Pnky knockdown. Heatmap representation of the differential expression of the overlapping gene set in shCtrl, shPnky, and shPtbp1 biological duplicate cultures. E) Venn Diagram demonstrating the overlap between exons showing differential usage (FDR < 0.01) upon Ptbp1 or Pnky knockdown. Heatmap representation of the differential usage of the overlapping gene set in shPnky and shPtbp1 cultures compared to control. F-H) RT-qPCR detection of expression of indicated genes normalized to Gapdh. Expression in each condition is shown relative to control (shCtrl-GFP, shCtrl-mCherry). Error bars are 95% confidence intervals from 3 separate cultures. See also Figure S4 and Tables S1-S3.