Abstract
Fusarium oxysporum f. sp. cubense (FOC) is an important soil-borne fungal pathogen causing devastating vascular wilt disease of banana plants and has become a great concern threatening banana production worldwide. However, little information is known about the molecular mechanisms that govern the expression of virulence determinants of this important fungal pathogen. In this study, we showed that null mutation of three mitogen-activated protein (MAP) kinase genes, designated as FoSlt2, FoMkk2 and FoBck1, respectively, led to substantial attenuation in fungal virulence on banana plants. Transcriptional analysis revealed that the MAP kinase signaling pathway plays a key role in regulation of the genes encoding production of chitin, peroxidase, beauvericin and fusaric acid. Biochemical analysis further confirmed the essential role of MAP kinases in modulating the production of fusaric acid, which was a crucial phytotoxin in accelerating development of Fusarium wilt symptoms in banana plants. Additionally, we found that the MAP kinase FoSlt2 was required for siderophore biosynthesis under iron-depletion conditions. Moreover, disruption of the MAP kinase genes resulted in abnormal hypha and increased sensitivity to Congo Red, Calcofluor White and H2O2. Taken together, these results depict the critical roles of MAP kinases in regulation of FOC physiology and virulence.
Introduction
Fusarium oxysporum f. sp. cubense (FOC) is an important soil-borne fungal pathogen causing vascular wilt disease of banana plants (also called Panama disease), which is the most important lethal disease of banana leading to serious crop losses in banana plantations [1]. The pathogen invades, colonizes and blocks the xylem vessels of the roots, and disrupts water and nutrient translocation resulting in severe plant wilting [2]. Typical symptoms of the disease include yellowing and wilting of the leaves, vascular discoloration inside the rhizome and pseudostem, and the infected plant eventually dies [1]. Four races of this pathogen have been described which attack different banana cultivars. Among them, race 4 is most devastating as it attacks much more banana cultivars than other races. Despite the importance of the disease caused by FOC, molecular mechanisms underlying pathogenicity and host infection of the fungus are poorly understood. So far, only two genes associated with the fungal virulence against banana plants have been characterized. Foatf1 encodes a bZIP transcription factor, which contributes to the full virulence of FOC by positively regulating the transcriptional expression of catalases to counteract the plant defenses mediated by reactive oxygen species [3]. FoOCH1 encodes a putative α-1, 6-mannosyltransferase, which plays a critical role in the maintenance of cell wall integrity and virulence [2].
Mitogen-activated protein (MAP) kinase cascades play crucial roles in transducing various extracellular signals and regulating growth and differentiation processes [4]. MAP kinase cascades include a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK) and a MAP kinase (MAPK) which is activated by dual phosphorylation of conserved threonine and tyrosine residues [5]. In the budding yeast Saccharomyces cerevisiae, a signaling pathway consisting of five distinct MAP kinases has been identified and shown to regulate mating, invasive growth, cell wall integrity, osmoregulation stress response, and ascospore formation [6]. In the human pathogen Aspergillus fumigatus, there are only three MAP kinases which regulate cell wall integrity, oxidative stress response, iron adaptation, adhesion, virulence and biosynthesis of secondary metabolism [7,8]. In Magnaporthe oryzae, similarly, three MAP kinases are involved in modulation of appressorium formation, pathogenicity, infectious growth, conidiation, cell wall integrity and oxidative stress response [9]. In Fusarium graminearum, the MAP kinase MGV1 is required for female fertility, heterokaryon formation and plant infection, but not conidiation [10]. In Alternaria alternate, the MAP kinase AaSLT2 governs conidiation, virulence and production of toxin and melanin [11]. In Coniothyrium minitans, deletion of the kinase genes CmBck1 and CmSlt2 involving in cell wall integrity affects conidiation and mycoparasitism, respectively [6]. The above findings highlight the importance of MAP kinase signaling mechanism in fungal physiology but also suggest that it may regulate different traits in different fungal species.
Fusarium oxysporium is an asexual fungal species consisting of many nonpathogens and pathogenic forms which infect plants, animals and human, respectively. Within the plant pathogens, over 150 pathogenic forms of F. oxysporium have been documented [12]. Among them, to our knowledge, only one MAP kinase gene Fmk1 in the tomato vascular wilt fungus Fusarium oxysporum f. sp. lycopersici was characterized and shown to play a key role in modulation of infectious growth, root penetration and pathogenesis [13]. Given the multiple evolutionary origins of F. oxysporum [14], it would be interesting to determine and compare the roles of the MAP kinase signaling pathway in various F. oxysporium species.
The objectives of this study are to investigate the roles of MAP kinases in FOC physiology and virulence. Based on bioinformatics analysis, the mutants of three MAP kinase genes, i.e., FoSlt2, FoMkk2 and FoBck1, were generated and characterized. The results from this study indicate that the MAP kinases are involved in regulation of a range of physiological traits and virulence determinants, including cell wall integrity, anti-oxidative mechanisms, and production of fusaric acid, which is a crucial phytotoxin in accelerating the development of Fusarium wilt in banana plants [15]. Additionally, we showed that the MAP kinase FoSlt2 was required for siderophore biosynthesis when FOC was grown under iron-limited conditions. Furthermore, we found that null mutation of three MAP kinase genes led to drastic attenuation in virulence when compared with their parental wild type fungal pathogen.
Results
In silico analysis of the MAP kinase genes FoSlt2, FoMkk2 and FoBck1
Considering the various role of MAP kinases in other fungal organisms, we set to search for the presence of their homologues in F. oxysporum f. sp. cubense (FOC) tropical race 4 strain XJZ2, which was isolated from diseased banana in Guangdong Province, China [2]. As the genome sequence of strain XJZ2 is not yet available, we firstly search the genome sequence of FOC tropical race 4 strain II5 (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html) using the coding sequences of the MAP kinase (MAPK) gene MGV1 of F. graminearum [10], the MAP kinase kinase (MAPKK) gene Mkk2 and the MAP kinase kinase kinase (MAPKKK) gene Bck1 from A. fumigatus [8] as query sequences. The search identified three orthologues at different locations of the strain II5 genome. We then amplified the three orthologous genes from FOC strain XJZ2 by using the primers designed according to the corresponding DNA sequences of strain II5. DNA sequence analysis showed that both FOC strains contain identical coding sequences for the three MAP kinase genes. Among them, FOIG_09199 herewith named FoSlt2 contains an open reading frame (ORF) of 1545 bp with four intervening introns (111bp, 71bp, 50bp, 56bp) and encodes a peptide of 419 amino acids that shows 97% identity to the MAPK MGV1 of F. graminearum. FOIG_05686 named FoMkk2 contains an ORF of 1733 bp with three intervening introns (55bp, 48bp, 49bp), and encodes a peptide of 527 amino acids that shows about 68% identity to the MAPKK Mkk2 of A. fumigatus; and FOIG_03241 designated as FoBck1 contains an ORF of 5721 bp with three intervening introns (55bp, 198bp, 50bp), and encodes a peptide of 1806 amino acids that shares about 49% identity to the MAPKKK Bck1 of A. fumigatus. Domain analysis showed that FoSlt2, FoMkk2 and FoBck1 all contain the conserved catalytic domain of the Serine/Threonine Kinases (S1 Fig).
Phylogenetic analysis showed that FoSlt2 clustered with F. oxysporum Fo5176 hypothetical protein FOXB_06615. FoMkk2 clustered with Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE7/MKK protein kinase, Fusarium oxysporum Fo5176 hypothetical protein FOXB_03604, Fusarium oxysporum f. sp. melonis 26406 STE/STE7/MKK protein kinase, Fusarium fujikuroi IMI 58289 probable MAP kinase kinase and Fusarium verticillioides 7600 STE/STE7/MKK protein kinase. FoBck1 clustered with Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE11/BCK1 protein kinase and Fusarium oxysporum f. sp. lycopersici MN25 STE/STE11/BCK1 protein kinase. The results showed that the FOC MAP kinases are highly conserved in Fusarium species, but they are less similar to their counterparts in other fungal species, especially the MAP kinases from A. fumigatus (S2 Fig).
Mutation of MAP kinase genes affects FOC hyphal growth but has no effect on fungal conidiation
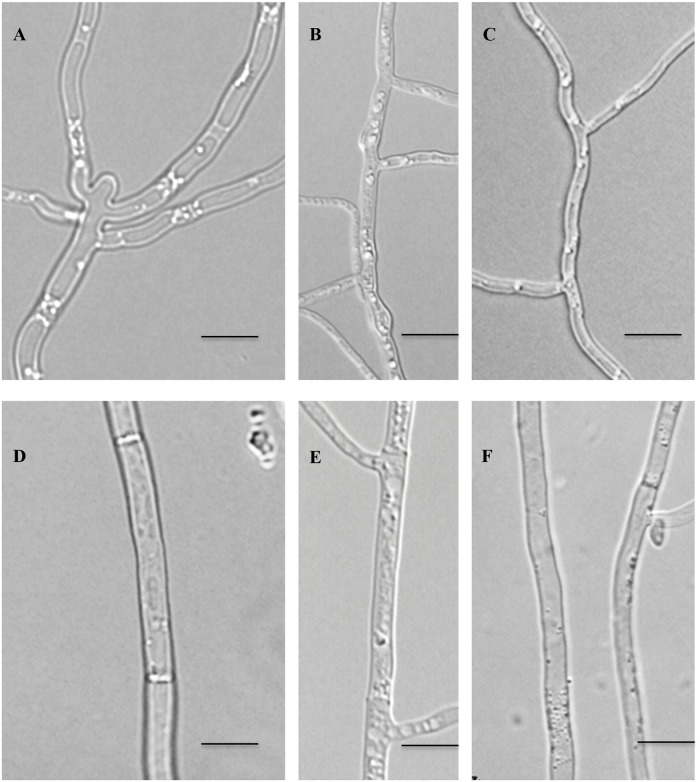
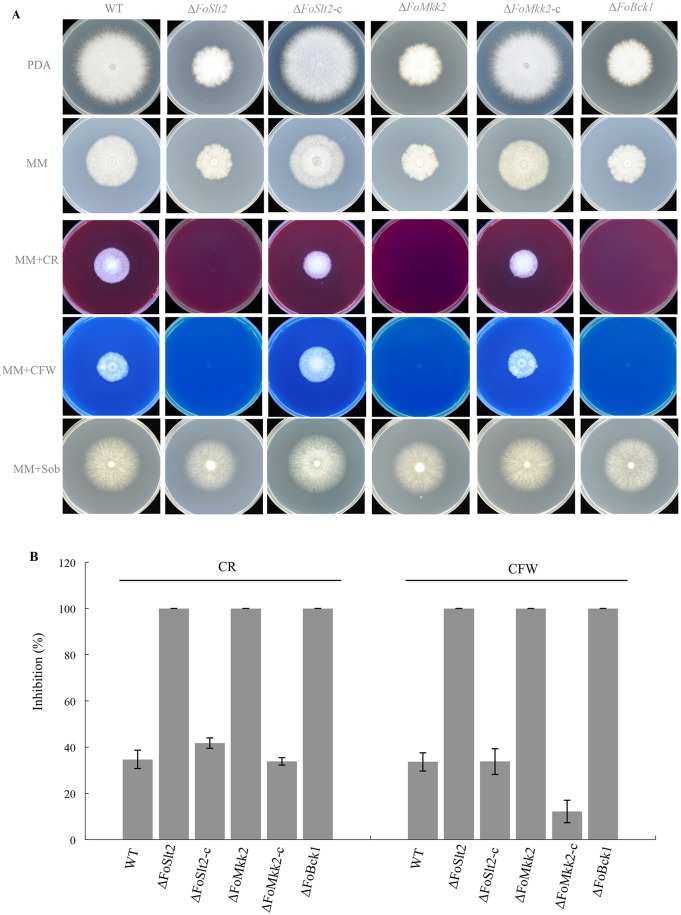
The high similarity of MAP kinase genes among Fusarium species facilitated generation of corresponding mutants in FOC race 4 strain XJZ2. To generate the knockout mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, the upstream and downstream sequences of the MAP kinase genes FoSlt2 (Slt2), FoMkk2 (Mkk2), and FoBck1 (Bck1) were amplified by PCR (S3 Fig, S1 Table), and fused with the hph gene encoding hygromycin resistance, respectively (S3 Fig). Thus these three MAP kinase genes were replaced separately by hph through homologous recombination. The complemented strains ΔFoSlt2-c and ΔFoMkk2-c were generated by cloning of the wild type FoSlt2 and FoMkk2 under the control of native promoter in the vector pMD18-T before inserting a zeocin resistance cassette (S3 Fig). The knockout mutants and complemented strains were selected in the medium containing appropriate antibiotics, and validated by PCR, Southern blot analysis and quantitative real-time PCR (S3–S6 Figs, S1 Table). Bright field microscopy revealed that the three mutants had flexuous hyphal structures and contained more branches compared with WT (Fig 1A–1D), which were restored in the complemented strains ΔFoSlt2-c and ΔFoMkk2-c (Fig 1E and 1F). On PDA or MM plates, the mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 showed similar morphologies with their colonies being smaller and more compact than that of WT (Fig 2A). Additionally, while WT produced abundant aerial hyphae on PDA plates, the three mutants produced fewer and shorter aerial hyphae (Fig 2A). The hyphal growth rate of the three mutants was assayed on MM plates with WT as a control. The results showed that the hyphal growth rate of the three mutants was lower than that of WT (Table 1). Moreover, the colony morphology of the mutants could be restored to the WT levels on MM plates supplemented with 1.2 M sorbitol (Fig 2A), indicating that the integrity of the cell wall is to some extent complemented by osmotic stabilizer.
Fig 1. Hypha morphology of WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, and complemented strains ΔFoSlt2-c and ΔFoMkk2-c.
Strains were incubated at 28°C for 4 days on PDA plates. A: ΔFoSlt2, B: ΔFoMkk2, C: ΔFoBck1, D: WT, E: ΔFoSlt2-c, F: ΔFoMkk2-c. Bars: 10μm.
Fig 2. Effects of FoSlt2, FoMkk2 and FoBck1 genes on cell wall integrity of FOC.
(A) Colony morphology of the indicated strains grown on PDA, minimal medium (MM), MM supplemented with Congo Red (CR, 40 μg/mL), Calcofluor White (CFW, 40 μg/mL), or sorbitol (Sob, 1.2 M) incubated at 28°C for 6 days on MM plates. (B) Inhibition of the radiated growth of the indicated strains grown on the MM plates. Error bars indicate the standard error from three replicates.
Table 1. Comparison of hyphal growth rate, conidiation and biomass among mutants and WT of FOC.
| Strains | Growth rate (mm/d) a | Conidiation (×106 conidia/mL) | Dry weight (g) d | |
|---|---|---|---|---|
| Static culture b | Shaken culture c | |||
| WT | 11.17 ± 0.01 A e | 2.67 ± 0.33 A | 25.00 ± 7.64 A | 3.17 ± 0.04 A |
| ΔFoSlt2 | 6.23 ± 0.01 C | 3.00 ± 0.88 A | 16.67 ± 1.67 A | 2.97 ± 0.02 A |
| ΔFoSlt2-c | 11.60 ± 0.02 A | 2.50 ± 0.58 A | 18.33 ± 3.99 A | 3.16 ± 0.16 A |
| ΔFoMkk2 | 6.60 ± 0.01 C | 4.00 ± 0.76 A | 20.00 ± 2.89 A | 3.26 ± 0.02 A |
| ΔFoMkk2-c | 11.63 ± 0.01 A | 3.33 ± 0.60 A | 26.67 ± 1.67 A | 3.14 ± 0.09 A |
| ΔFoBck1 | 7.03 ± 0.01 B | 3.17 ± 0.33 A | 20.00 ± 5.78 A | 3.07 ± 0.15 A |
a Growth rate was detected by measuring the colony diameter of cultures incubated on PDA plates after 6 days at 28°C.
b, c Conidia produced by static and shaking liquid cultures in YPG for 48 h at 28°C and counted with haematocytometer.
d Dry weight of fungal biomass grown in Czapek dox medium.
e Different letters within a column indicate statistically significant differences (P = 0.05). Error bars indicate the standard error from three replicates.
Interestingly, although mutation of these three MAP kinase genes showed substantial effect on the fungal hyphal structure and colony morphology, the conidia produced by the three mutants were similar to WT with normal size and morphology in YPG liquid medium under either static or shaken culture (data not shown). We also found the biomass of the three mutants was not significantly different from WT and complemented strains ΔFoSlt2-c and ΔFoMkk2-c (Table 1). Thus, these three MAP kinases may be important for FOC development and elongation of aerial hyphae.
The MAP kinases govern cell wall integrity
The sensitivities of WT, mutants and complemented strains to cell wall inhibitors Congo red and Calcofluor white were assessed. The results showed that three mutants were more sensitive to Congo red and Calcofluor white than WT, and the mutants phenotypes could be rescued by in trans expression of the corresponding wild type genes in complemented strains (Fig 2A). Quantitative analysis indicated that addition of the two cell wall inhibitors resulted in much more severe inhibition on growth of the MAP kinases mutants compared with WT and the complemented strains (Fig 2B).
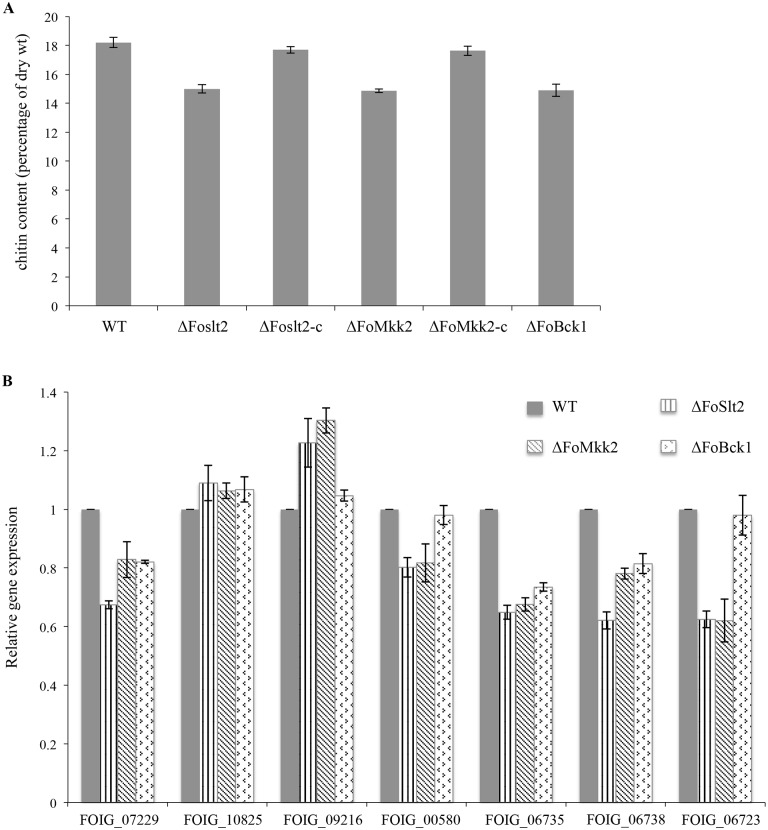
Chitin is one of the major components of the Fusarium oxysporum cell wall [16]. The mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 showed lower chitin contents than WT and the complemented strains ΔFoSlt2-c and ΔFoMkk2-c (Fig 3A). Given that synthesis of chitin is dependent on the activity of chitin synthase [17], we analyzed the expression levels of seven chitin synthases with quantitative real-time PCR. As expected, the results showed that except for FOIG_00580 and FOIG_06723 in the mutant ΔFoBck1, the expression levels of five chitin synthase genes including FOIG_07229, FOIG_00580, FOIG_06735, FOIG_06738 and FOIG_06723 were reduced significantly in the three mutants compared with the WT control (Fig 3B).
Fig 3. Quantification of chitin and expression of chitin synthase genes.
(A) Chitin content in WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, complemented strains ΔFoSlt2-c and ΔFoMkk2-c. (B) Quantitative real-time PCR analysis of chitin synthase in WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1. The seven chitin synthase are FOIG_07229, FOIG_10825, FOIG_09216, FOIG_00580, FOIG_06735, FOIG_06738 and FOIG_06723. Error bars indicate the standard error from three replicates.
Disruption of the MAP kinase genes affects the fungal sensitivity to hydrogen peroxide
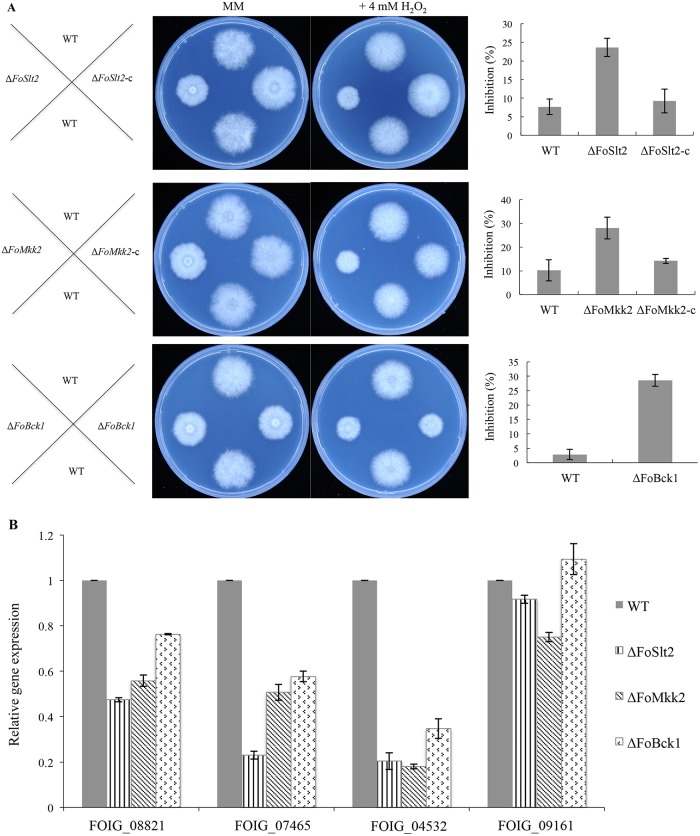
The sensitivities of WT, three MAP kinase mutants and the complemented strains ΔFoSlt2-c and ΔFoMkk2-c to oxidative stress were tested. The results showed that three mutants were more sensitive to 4 mM H2O2 than WT and the corresponding complemented strains (Fig 4A). Moreover, quantitative real-time PCR analysis showed that except for FOIG_09161 in mutant ΔFoBck1, the four peroxidase synthase genes were down-regulated in the three mutants compared with WT (Fig 4B). Collectively, these findings indicate that the MAP kinases may play a vital role in regulation of the degradation of extracellular reactive oxygen species (ROS). Generation of ROS is a well-known hallmark event in host plant defense mechanisms [18].
Fig 4. The deletion mutants were sensitive to H2O2.
(A) Middle panels of figures show colony morphology of the WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, complemented strains ΔFoSlt2-c and ΔFoMkk2-c were incubated on MM plates supplemented with 4 mM H2O2 at 28°C for 3.5 days. Right panels of figures show inhibition of the radiated growth of the indicated strains grown on the MM plates with the same order as the middle figures. (B) Quantitative real-time PCR analysis of peroxidase synthase genes in the WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1. The four peroxidase synthase genes are FOIG_08821, FOIG_07465, FOIG_04532 and FOIG_09161. Error bars indicate the standard error from three replicates.
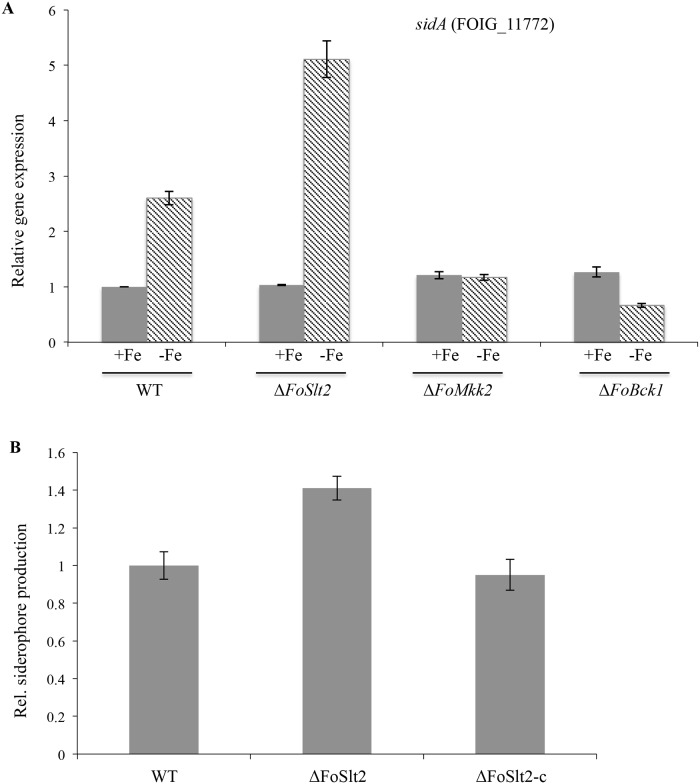
The FoSlt2 gene is involved in siderophore biosynthesis
FOIG_11772 encodes a predicted orthologue of the L-ornithine N (5)- monooxygenase encoded by the sidA gene of Aspergillus nidulans [19]. To test whether the expression of this siderophore biosynthetic gene is influenced by disruption of the MAP kinase genes, we grew WT and three MAP kinases mutants in iron-poor and-replete conditions. Quantitative real-time PCR analysis showed that the siderophore biosynthetic gene sidA was sharply upregulated in mutant ΔFoSlt2 (2-fold) but not in ΔFoMkk2 and ΔFoBck1 during iron-poor conditions (Fig 5A). In line with quantitative real-time PCR data, a chrome azurol S (CAS) assay detected about a 1.4-fold increase in siderophore level in mutant ΔFoSlt2 in comparison with the WT (Fig 5B). These results demonstrate that FoSlt2 is involved in the regulation of siderophore biosynthesis in FOC during iron-poor conditions.
Fig 5. Determination of siderophore biosynthesis in mutant ΔFoSlt2 during iron-poor conditions.
(A) The siderophore biosynthetic gene sidA was upregulated in mutant ΔFoSlt2 during iron-poor conditions, normalized to WT using quantitative real-time PCR. (B) CAS assay showed increased level of secreted siderophores in mutant ΔFoSlt2 during iron-poor conditions, normalized to WT. The indicated strains were incubated for 5 days and the supernatants were analyzed for siderophore content. Error bars indicate the standard error from three replicates.
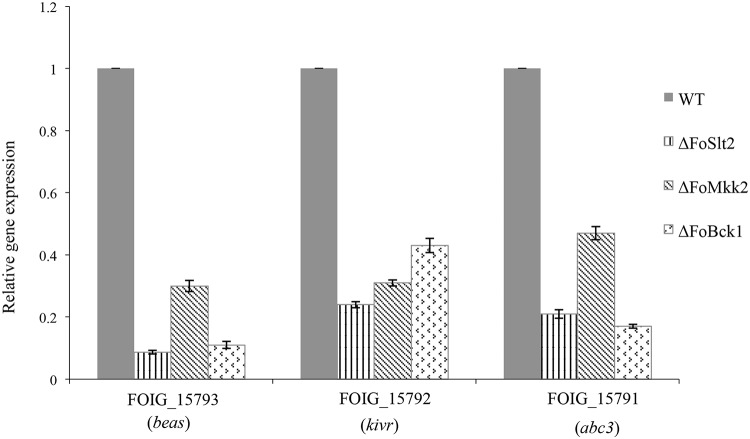
The MAP kinases are involved in regulation of the transcription of beauvericin biosynthetic genes
Beauvericin is a cyclohexadepsipeptide mycotoxin which shows insecticidal properties and can induce apoptosis in mammalian cells [20]. To understand the putative effect of the MAP kinases on mycotoxin production, we identified three genes (FOIG_15793, FOIG_15792 and FOIG_15791) in the FOC4 genome which are the orthologues of the beauvericin biosynthetic genes from F. oxysporum f. sp lycopersici race 2. FOIG_15793 encodes a predicted orthologue of enniatin and beauvericin synthetases beas, FOIG_15792 encodes a putative orthologue of 2-ketoisovalerate reductase kivr and FOIG_15791 encodes a predicted orthologue of ABC multidrug transporter abc3 [21]. The three genes FOIG_15793, FOIG_15792 and FOIG_15791 show over 97% identity with their counterparts in F. oxysporum f. sp lycopersici race 2 at amino acids level.
Quantitative real-time PCR analysis confirmed the role of the three MAP kinases in regulation of the transcriptional expression of the beauvericin biosynthetic genes. The expression levels of beas, kivr and abc3 genes were significantly reduced in mutants ΔFoSlt2 (12-fold, 4-fold and 5-fold, respectively), ΔFoMkk2 (3-fold, 3-fold and 2-fold, respectively) and ΔFoBck1 (9-fold, 2-fold and 6-fold, respectively) than those of WT, respectively (Fig 6).
Fig 6. The MAP kinases affect the transcription of beauvericin biosynthetic genes.
Gene expression levels are represented relative to WT using quantitative real-time PCR. Error bars indicate the standard error from three replicates.
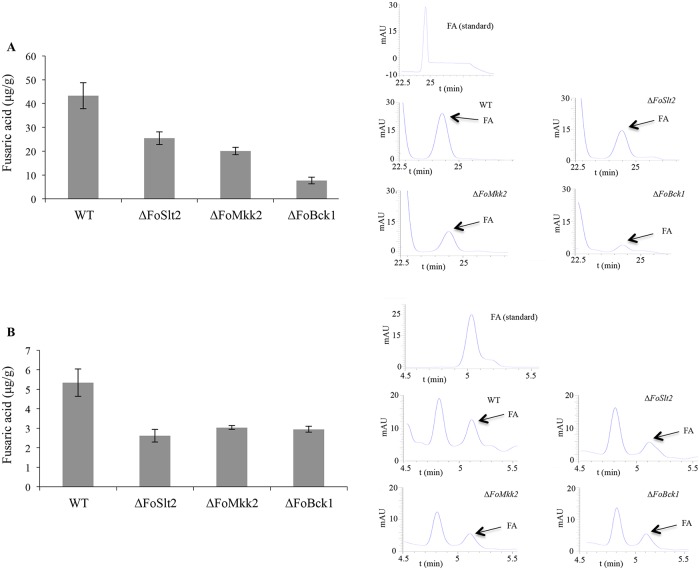
The MAP kinases are involved in fusaric acid biosynthesis
Fusaric acid is a mycotoxin with low to moderate toxicity to animals and humans, but with high phytotoxic properties [22], and is thought to cause the severity of F. oxysporum-induced vascular wilt, damping-off and root rot diseases of numerous vegetable crops [23]. By searching the homologous genes of the fusaric acid biosynthetic genes of F. verticillioides, we identified five genes (FUB1 to FUB5) in the genome of FOC. Among them, FOIG_16450 (FUB1) encodes an orthologue of PKS from F. verticillioides, which catalyzes condensation of three acetate units to form a fully reduced 6-carbon polyketide chain, FOIG_16452 (FUB3) encodes a putative orthologue of F. verticillioides amino acid kinase, which likely plays a critical role in assimilating a nitrogen from glutamine or oxaloacetate to form fusaric acid, FOIG_16453 (FUB4) encodes a predicted hydrolase orthologue from F. verticillioides and FOIG_16454 (FUB5) is predicted to encode an acetyltransferase orthologue of F. verticillioides, which is responsible for the addition of a methyl group to the carboxylic acid moiety of fusaric acid to yield methyl fusarate [23]. FUB1-5 shows over 94% identity with their counterparts in F. verticillioides at amino acids level.
To determine the involvement of MAP kinases in the regulation of fusaric acid biosynthesis, we analyzed the expression levels of five genes involved in fusaric acid biosynthesis. The expression levels of fusaric acid biosynthetic genes (FUB1 to FUB5) were significantly reduced in mutants ΔFoSlt2 (100-fold, 11-fold, 10-fold, 25-fold and 50-fold, respectively), ΔFoMkk2 (100-fold, 13-fold, 10-fold, 20-fold and 33-fold, respectively) and ΔFoBck1 (50-fold, 6-fold, 6-fold, 17-fold and 25-fold, respectively) compared with that of WT, respectively. In the complemented strains ΔFoSlt2-c and ΔFoMkk2-c, expression levels of the five genes were completely or partially restored to the WT levels, respectively (Fig 7).
Fig 7. The MAP kinases regulate transcription of the fusaric acid gene cluster.
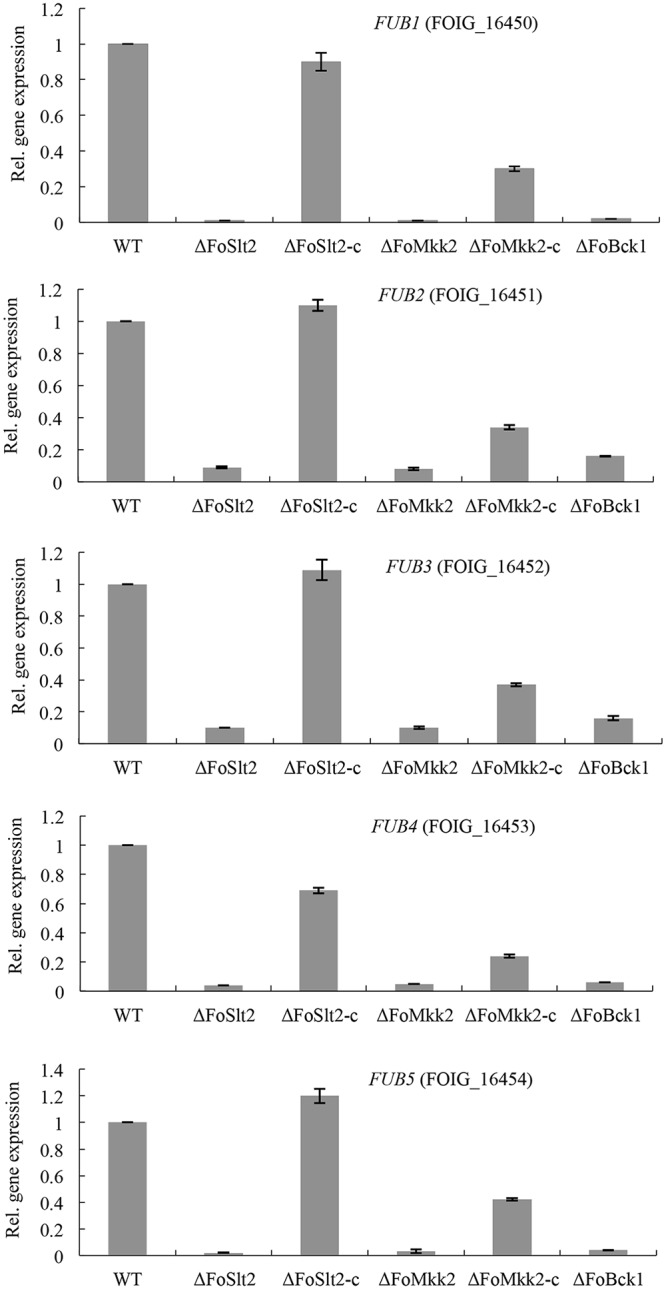
Gene expression levels are represented relative to WT using quantitative real-time PCR. Error bars indicate the standard error from three replicates.
We then determined the fusaric acid production in Czapek dox medium by WT and mutants by high performance liquid chromatography (HPLC). In agreement with the gene expression data, the production of fusaric acid was reduced in mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 compared with that of WT, respectively (Fig 8A).
Fig 8. Fusaric acid (FA) production was reduced in mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1.
(A) Fusaric acid production from cultures of the indicated strains in Czapek dox medium was analysed by High Performance Liquid Chromatography (HPLC). Fusaric acid levels are represented in μg g-1 mycelial dry weight. (B) Fusaric acid production from the crude mycotoxin extracted of the indicated strains in grains cultures was analysed with HPLC. Fusaric acid levels are represented in μg g-1 grains cultures dry weight. Error bars indicate the standard error from three replicates.
We further determined the fusaric acid production in grains culture by WT and the three mutants. The crude mycotoxin extracts from the indicated strains in grains cultures were also analyzed using HPLC. The result showed that fusaric acid production was similarly reduced in mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 (Fig 8B) compared with that of WT, respectively. We conclude that MAP kinases play a vital role in regulation of fusaric acid production.
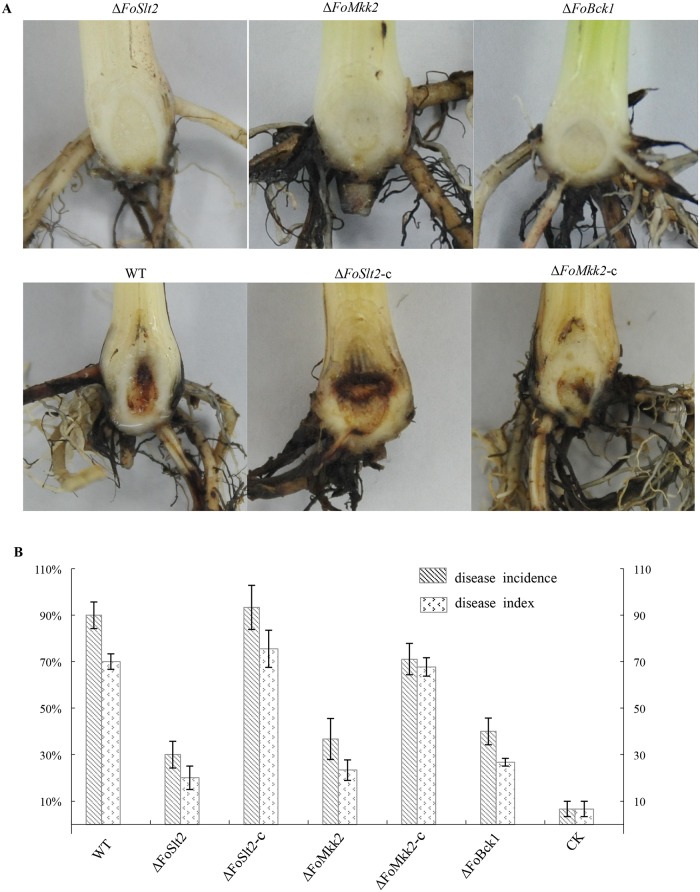
The MAP kinases are necessary for the fungal virulence on banana plants
Virulence tests showed that the mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 were unable to produce visible vascular discoloration in the corm of the banana plantlets (Fig 9A) and were significantly reduced in virulence on Cavendish banana compared to the WT according to disease incidence and disease index (Fig 9B), while the complemented strains ΔFoSlt2-c and ΔFoMkk2-c produced obvious internal disease symptoms of brown discoloration (Fig 9A) with virulence restored to the WT level (Fig 9B). These results indicate that MAP kinases are required for the full virulence of F. oxysporum f. sp. cubense.
Fig 9. Virulence assay of WT, mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, and complemented strains ΔFoSlt2-c and ΔFoMkk2-c.
(A) Disease symptom on corms of banana tissue plants were assayed after 40 days of inoculation with indicated strains and water as negative control. (B) Disease incidence and disease index indicated disease severity. Error bars indicate the standard error from three replicates.
Discussion
Mitogen-activated protein (MAP) kinases are crucial for fungal growth, conidiation, oxidative stress response, cell wall integrity, virulence and siderophore biosynthesis in plant and human pathogens, and are the targets for developing antifungal drugs [7,9]. In our study, three MAP kinase genes, FoSlt2, FoMkk2 and FoBck1 were identified and characterized in the soil-borne fungal pathogen FOC tropical race 4 strain XJZ2. Our data provide first evidence for the crucial role of the three MAP kinases in the maintenance of fungal development, cell wall integrity and virulence of FOC. In addition, the results from this study also unveil several new findings previously unreported in other plant fungal pathogens including the association of the MAP kinases with the FOC resistance to oxidative stress, the involvement of FoSlt2 in regulation of siderophore biosynthesis, and the critical role of MAP kinases in modulation of fusaric acid biosynthesis, which is a key virulence determinant.
Our results revealed that the hyphal growth rates of the mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 were reduced on solid media but not affected in liquid media. Our results are reminiscent to the mutants of the Slt2 orthologue in M. grisea and F. graminearum, which form fewer aerial hyphae than corresponding wild type controls on agar plates, while produced normal mycelial growth in liquid media [10,24]. Similarly, mutation of Slt2 orthologue also results in decreased hyphal growth in several other fungal species including plant pathogens A. alternate [11], Botrytis cinerea [25], and human pathogen A. fumigatus [8,26]. Interestingly, deletion of single FoSlt2, FoMkk2 and FoBck1 genes did not affect conidiation in FOC, which is similar to the MGV1 mutant (Slt2 orthologue) of F. graminearum [10]. On the contrary, deletion of the Slt2 orthologue in C. minitans [6], or A. alternata [11], and knockout of the Bck1 orthologue in M. oryzae [27] displayed reduced conidiation. Thus, the biological functions of the MAP kinases may vary considerably in different fungal species.
The mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 showed hypersensitivity to cell wall inhibitors including Congo red and Calcofluor white, which has also been reported in other fungal mutants, such as A. fumigatus ΔMpkA (Slt2 orthologue), ΔMkk2 and ΔBck1 [8], Candida albicans Δmkc1 (Slt2 orthologue) [28], A. alternate ΔAaSlt2 [11], and C. minitans ΔCmSlt2 [6]. Moreover, deletion of the single core element of MpkA MAP kinases resulted in aberrant hyphal growth in A. fumigatus [8]. We not only observed the aberrant hypha but also reduced production of chitin and reduced expression levels of chitin biosynthetic genes in the mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 compared with WT and complemented strains, which may explain why the three mutants become sensitive to cell wall inhibitors and cell wall degrading enzyme to release more protoplasts (data not shown) than the WT control. Previous transcriptome analysis revealed that the MAP kinase MpkA of A. fumigatus play a key role in regulation of the genes involved in cell wall remodeling, such as gel1 and yap2 encoding β-glucan synthesis [7,29]. Our results reconfirmed the importance of these three MAP kinases in the maintenance of cell wall integrity through regulation of the genes involved in biosynthesis of chitin.
Sensitivity assay revealed that the FOC mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 were hypersensitive to oxidative agent. The results were further consolidated by the findings that deletion of the MAP kinase genes led to decreased transcriptional expression of the four peroxidase synthase genes compared with WT. Our results apparently differ from the previous findings in other fungal species. The A. alternate mutant ΔAaSlt2 did not alter the cellular sensitivity to H2O2 [11] and A. fumigatus mutant ΔMpkA promoted tolerance against H2O2 [30]. To our knowledge, the results from this study for the first time established the link between MAP kinases and the anti-oxidation mechanisms in a plant fungal pathogen. These results also suggest that FOC may rely on MAP kinases to regulate anti-oxidation mechanisms, such as peroxidase to counteract the extracellular reactive oxygen species (ROS) produced by host plants during pathogen-host interactions.
We found the connection between the MAP kinase FoSlt2 and siderophore biosynthesis in plant pathogen. Iron is an essential nutrient for almost every organism for a variety of cellular processes but its excess is toxic to the cell. In fungi, a series of genes required for siderophore-mediated iron uptake are upregulated during iron-poor condition, while iron-consuming pathways are rapidly down-regulated to cope with limited availability of iron [31]. Siderophores are a class of small molecules that are produced by organisms to chelate iron [32]. F. oxysporum excretes four major siderophores, including extracellular fusarinine C (FSC) and malonichrome, and intracellular ferricrocin (FC) and ferrichrome C [31]. The siderophores assembled by NRPS systems are strictly regulated by the availability of iron [7,31]. The loss of siderophores due to deficiency in the ornithine monooxygenase SidA, or the iron regulator HapX or the NRPS gene NPS6 reduced virulence in human and plant pathogens, which may be the reason why the mammalian or plant hosts sequester iron from invading microorganisms [31–35]. The experimental results showed that deletion of FoSlt2 but not FoMkk2 and FoBck1 in FOC led to increased siderophore production by enhancing sidA expression during iron-poor conditions. Thus far, the Slt2 orthologue MpkA has been suggested to control siderophore biosynthesis by acting as a repressor of sidA in the human pathogen A. fumigatus [7]. Siderophores commonly contains ornithine derivatives in the peptide backbones, and SidA performs first enzymatic step in preparation of these derivatives using ornithine as the substrate. Ornithine is also a key amino acid precursor for polyamines formation, and polyamines are important for fungal growth and development [7,33]. In A. fumigatus, interestingly, the polyamine level in the wild type strain decreased during iron starvation while it remained constant in the ΔMpkA mutant. This suggests that the MAP kinase MpkA acts as repressor of ornithine monooxygenase SidA to divert available ornithine in production of polyamines for essential survival processes under iron depletion conditions [7]. While the mechanisms of regulation remain vague, the above information from A. fumigatus may provide useful clues for further investigation on how FoSlt2 modulates the transcriptional expression of siderophore genes in FOC.
We also found that the MAP kinases were involved in fusaric acid biosynthesis. Members of the genus Fusarium are known to produce a diversity of toxic secondary metabolites, such as fumonisins, trichothecenes, fusaric acid and beauvericin, which are threats to plant growth, animal and human health [23,36–38]. Fusaric aicd is a well-known nonspecific toxin produced by all Fusarium species, which may alter cell growth, mitochondrial activity and membrane permeability, and even though plays a direct role in fungal pathogenicity [15]. Previous studies revealed that production of fusaric acid is encoded by fusaric acid biosynthetic gene cluster (FUB1-FUB5). Deletion of FUB1 led to loss of fusaric aicd production in F. verticillioides [23]. In F. fujikuroi, deletion of either FUB1 or FUB4 also caused a total loss of fusaric acid production, while the mutants ΔFUB2, ΔFUB3 and ΔFUB5 were still able to produce fusaric acid with reduced amounts compared with the wild type strain [22]. Expression of fusaric aicd genes is influenced by nitrogen and pH changes with nitrogen-dependent expression positively regulated by the nitrogen-responsive GATA transcription factor AreB and pH-dependent regulation mediated by the transcription factor PacC [22]. It is intriguing to test whether these MAP kinases regulate fusaric acid synthesis through AreB or PacC or other regulators.
In addition, we found that the MAP kinases regulated the transcription of beauvericin biosynthetic genes. Beauvericin is toxic cyclic hexadepsipeptide and a virulence factor on mammals and plants and its production has been reported in several plant pathogenic Fusarium species [20,36]. Previous studies revealed that reduced virulence of mutants ΔvelA, ΔvelB and ΔlaeA of F. oxysporum on mice and on tomato plants was accompanied by decreased levels of beauvericin [21]. However, little information is available on how beauvericin production is regulated in FOC. The findings from this study that the mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1 showed decreased expression of fusaric acid and beauvericin biosynthetic genes, and reduced production of fusaric acid present new members to the list of MAP kinases regulated toxic secondary metabolites in fungal pathogens.
In summary, this study has characterized three MAP kinase genes of FOC, including the MAPK gene FoSlt2, the MAPKK gene FoMkk2 and the MAPKKK gene FoBck1. Consistent with previous notion that MAPK-MAPKK-MAPKKK from a signaling cascade, the deletion mutants of three MAP kinase genes showed almost identical phenotypes except that only FoSlt2 is involved in siderophore biosynthesis. Given the important roles of MAP kinases in regulation of various physiological traits and virulence determinants in FOC, including cell wall integrity, anti-oxidation enzymes, and biosynthetic genes encoding siderophore, phytotoxins fusaric acid and beauvericin, it is not surprisingly that null mutation of any of the three MAP kinase genes could lead to substantially attenuated fungal virulence. In summary, we have conducted detailed investigation on the biological functions regulated by MAP kinases in FOC, and our results showed that the MAP kinases play vital roles in regulation of various physiological traits and virulence determinants in FOC. A further understanding on this important signaling pathway and regulatory mechanisms may facilitate the development of new control strategies against this devastating fungal pathogen.
Materials and Methods
Fungal strains, media and culture conditions
F. oxysporum f. sp. cubense (FOC) race 4 strain XJZ2 was used as wild type (WT) for fungal transformation and gene knockout experiments as described [2]. Fungal strains were stored as microconidial suspensions at -80°C with 20% (v/v) glycerol. Potato dextrose agar (PDA), MM [39], yeast extract peptone dextrose (YPD) liquid medium and YPG liquid medium were used for characterization of vegetative growth and asexual development. To determine fungal growth rates, freshly obtained microconidia (2 μL; 108 microconidia per milliliter) were spotted onto MM plates, and MM plates were used as control. Growth rate was assayed by measuring the colony diameters after incubation for 6 days at 28°C. Conidiation was quantified in static or shaken liquid cultures as described [40].
Construction of plasmids and fungal transformation
For disruption of the targeted genes, plasmids pCT74-FoSlt2-KO, pCT74-FoMkk2-KO and pCT74-FoBck1-KO were generated. The upstream and downstream fragments of corresponding targeted genes were amplified from genomic DNA of WT and were sequenced. For FoSlt2 gene, upstream fragment using primer pair F1/R1, downstream fragment using primer pair F2/R2. For FoMkk2 gene, upstream fragment using primer pair F4/R4, downstream fragment using primer pair F5/R5. For FoBck1 gene, upstream fragment using primer pair F6/R6, downstream fragment using primer pair F7/R7. The SpeI-ClaI double-digested FoSlt2, FoMkk2 and FoBck1 upstream fragments were inserted into the same sites on pCT74 vector [41] to generate plasmids pCT74-FoSlt2-up, pCT74-FoMkk2-up and pCT74-FoBck1-up, respectively. Then the ApaI-KpnI double-digested FoSlt2, FoBck1 downstream fragments, and the XhoI-BglII double-digested (blunt-ended) FoMkk2 downstream fragment were inserted into the same clone sites of corresponding vectors pCT74-FoSlt2-up and pCT74-FoBck1-up, and XhoI site (blunt-ended) of pCT74-FoMkk2-up vectors to generate plasmids pCT74-FoSlt2-KO, pCT74-FoBck1-KO and pCT74-FoMkk2-KO, respectively. The plasmids pCT74-FoSlt2-KO, pCT74-FoMkk2-KO and pCT74-FoBck1-KO were linearized with XbaI, PvuII and SacII, and then introduced into protoplasts of WT, respectively.
For complementation of the deletion mutants, plasmids pMD18-FoSlt2-COM and pMD18-FoMkk2-COM were generated. Complemented entire genes including the promoter region, the coding region and the terminator region were amplified from genomic DNA of WT and were sequenced, FoSlt2 gene fragment of 3.6-kb using primer pair HB-F1/HB-R1, FoMkk2 gene fragment of 3.9-kb using primer pair HB-F2 /HB-R2, and then cloned into the pMD18-T vector (Takara, Dalian, China), giving pMD18-FoSlt2 and pMD18-FoMkk2, respectively. Subsequently, a 1.2-kb BamHI-digested (blunt-ended) zeocin resistance cassette from plasmid pZGR1 was inserted into XbaI and SphI site (blunt-ended) of pMD18-FoSlt2 and pMD18-FoMkk2, resulting in plasmids pMD18-FoSlt2-COM and pMD18-FoMkk2-COM, respectively. The plasmids pMD18-FoSlt2-COM and pMD18-FoMkk2-COM were linearized with EcoRI and SalI, and then introduced into protoplasts of the ΔFoSlt2 and ΔFoMkk2 mutants, respectively.
Protoplasts of FOC were produced as described [42]. The fungal transformation according to a protocol described previously [43]. Colonies appeared after 4 days and were transferred on PDA plate containing 50 μg/mL of hygromycin B or 50 μg/mL zeocin, and were incubated at 28°C. Transformants were identified by PCR and southern blot analysis [2].
RNA manipulation and quantitative real-time PCR analysis
For analysis of gene expression, RNA of WT and mutants was extracted and quantitative real-time PCR was conducted as described previously [2]. For analysis of gene expression influenced by 1mM of FeSO4, we grew WT and mutants in MM during iron-poor and-replete conditions as described previously [31]. Transcript levels were calculated by comparative ΔCt and normalized to the endogenous control actin. Target gene expression values in mutants are presented as values relative to the expression in the WT.
Cell wall sensitivity assay
To test the sensitivities of the WT, mutants ΔFoSlt2, ΔFoMkk2, and ΔFoBck1, complemented strains ΔFoSlt2-c and ΔFoMkk2-c to cell wall inhibitors or H2O2, freshly obtained microconidia (2 μL; 108 microconidia per milliliter) were spotted onto MM plates and MM plates supplemented with sorbitol (Sob, 1.2 M), or Congo red (CR, 40 μg/mL), or calcofluor white (CFW, 40 μg/mL), or H2O2 (4 mM), respectively, and MM plates were used as control. Cell wall sensitivity to the chemicals mentioned above was assayed by measuring the colony diameters after incubation for 6 days (for cell wall inhibitor) or 3.5 days (for H2O2) at 28°C [8,44].
Chitin determination
Fungal cell wall was isolated as described [11]. Chitin was determined by measuring the acid-released glucosamine from chitin using p-dimethylaminobenzaldehyde as a chromogen. The absorbance at 520 nm was measured and the quantity of glucosamine was calculated by reference to a standard curve of 0–250 μg of glucosamine [45,46].
CAS assay
The measurement of siderophore production of the WT and mutant ΔFoSlt2 was carried out using CAS (chrome azurol S) assay as described previously [31]. Quantity of siderophores was calculated based on the standard curve of desferrioxamine.
Phylogenetic and bioinformatics analysis
The orthologous protein sequences of FoSlt2, FoMkk2 and FoBck1 were downloaded from NCBI GenBank database and Fusarium Comparative Database (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html). Clustal X version 2.0 [47] was used to align FoSlt2, FoMkk2 and FoBck1 and their orthologous, and the MEGA version 5.2 [48] was used to produce the phylogenetic tree. Bootstrap values were expressed as a percentage of 1000 replicates. ORF and exon/intron positions were identified by comparing genomic and cDNA sequences. Functional domains of FoSlt2, FoMkk2 and FoBck1 genes were determined using Conserved Domain Architecture Retrieval Tool (CDART) available from NCBI database.
Fusaric acid quantification
For liquid medium fermentation, 107 microconidia of WT and mutants ΔFoSlt2, ΔFoMkk2, and ΔFoBck1 were inoculated in 250 mL Czapek Dox medium and incubated at 28°C on a rotary shaker at 170 rpm for 9 days, the cultures were filtrated with Waterman filters to exclude mycelium and conidia, and crude mycotoxion was extracted as described [15]. An Agilent 1260 RP-HPLC system with an Agilent HC-C18 column (4.6 x 250 mm) was employed to analysis fusaric acid (FA). Elution was carried out using a mobile phase comprising 20% methanol, 48% double distilled water and 32% H3PO4 (0.43%) for 40 min with a UV detector at 280 nm, the flow rate was 1 mL/min. Before injection, the samples were filtrated through 0.45-μm filters. For solid medium fermentation, 250 mL glass bottles containing 50 g of grains mixture (wheat/barley/oats, 1/1/1) and 25mL of distilled water were stopped with cotton plugs and autoclaved at 121°C for 30 min, glass bottles were inoculated with 108 microconidia of WT and the three mutants and cultured at 28°C, respectively. After 23 days, the grains cultures were collected and dried. The crude mycotoxion was extracted from 2 g grains cultures with 10 mL acetonitrile/water/acetic acid (79/20/1, v/v/v) by sonication for 20 min, and then supernatant and residue were separated. Fusaric acid was analysed as above described with some modifications, 60% methanol, 5% double distilled water and 35% H3PO4 (0.43%) was used for mobile phase.
Virulence assay
The virulence of the three gene deletion mutants was tested on tissue culture-derived banana plantlets (Cavendish banana, AAA) at the 4–5 leaf stage. Banana root inoculation assays were performed as described [2]. Disease symptoms were assessed 40 days after inoculation. Thirty plantlets were used for each treatment.
Data analysis
Statistical analysis of the data was carried out using SPSS 21.0 software, with a completely randomized analysis of the variances (P = 0.05). Tukey’s honest significant difference (HSD) test was used for comparison of the means.
Accession Numbers
Sequence data can be found in the Fusarium Comparative Genome database under the following accession numbers: FoSlt2, FOIG_09199; FoMkk2, FOIG_05686; FoBck1, FOIG_03241; actin, FOIG_02823; seven chitin synthase genes, FOIG_07229, FOIG_10825, FOIG_09216, FOIG_00580, FOIG_06735, FOIG_06738 and FOIG_06723; four peroxidase synthase genes, FOIG_08821, FOIG_07465, FOIG_04532 and FOIG_09161; beas, FOIG_15793; kivr, FOIG_15792; abc3, FOIG_15791; sidA, FOIG_11772; FUB1, FOIG_16450; FUB2, FOIG_16451; FUB3, FOIG_16452; FUB4, FOIG_16453; FUB5, FOIG_16454.
Supporting Information
(TIF)
The GenBank accession numbers are Magnaporthe grisea Mps1 (AF020316), Aspergillus fumigatus MpkA (XM_746366, AFUA_4G13720), Colletotrichum lagenarium Maf1 (AY064246), Fusarium graminearum Mgv1 (AF492766), Aspergillus nidulans MpkA (U59214), Fusarium fujikuroi IMI 58289 MAP kinase (CCT68358), Fusarium verticillioides 7600 CMGC/MAPK/ERK1 protein kinase (EWG40764), Fusarium proliferatum putative MAP kinase (ABD67163), Fusarium oxysporum Fo5176 hypothetical protein FOXB_06615 (EGU82812), Aspergillus fumigatus Mkk2 (XM_745237, AFUA_1G05800), Neurospora crassa OR74A MAP kinase kinase (EAA28074), Fusarium oxysporum Fo5176 hypothetical protein FOXB_03604 (EGU85756), Fusarium oxysporum f. sp. melonis 26406 STE/STE7/MKK protein kinase (EXK41584), Fusarium fujikuroi IMI 58289 probable MAP kinase kinase (CCT66161), Fusarium verticillioides 7600 STE/STE7/MKK protein kinase (EWG44090), Magnaporthe oryzae 70–15 STE/STE7 protein kinase (XP_003717079), Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE7/MKK protein kinase (EXM04167), Magnaporthe grisea Mck1 (XP_368361), Aspergillus fumigatus Bck1 (XM_749418; AFUA_3G11080), Coniothyrium minitans Bck1 (JF951364), Fusarium oxysporum f. sp. lycopersici MN25 STE/STE11/BCK1 protein kinase (EWZ92886), Fusarium verticillioides 7600 STE/STE11/BCK1 protein kinase (EWG43584), Fusarium graminearum PH-1 hypothetical protein FGSG_06326 (ESU12405), Fusarium fujikuroi IMI 58289 bck1-like MAPKKK (CCT67356), Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE11/BCK1 protein kinase (EXM06456). All protein sequences were aligned using Clustal × 2.0. Aligned sequences were analyzed by Poisson model method in MEGA 5.2. Bootstrap values were calculated from 1000 bootstrap replicates.
(TIF)
(A) Strategic map of gene replacement and complementation construct, and restriction map of the FoSlt2 genomic region. (B) Strategic map of gene replacement and complementation construct, and restriction map of the FoMkk2 genomic region. (C) Strategic map of gene replacement and restriction map of the FoBck1 genomic region. The relative positions of the primers (short arrows) used for amplification of the linear DNA fragment employed for gene replacement, identification, quantitative real-time PCR and Southern blot analysis are indicated. H, HindIII; N, NcoI; B, BamHI; S, ScaI.
(TIF)
For the FoSlt2 gene, three FoSlt2 deletion mutants were identified by PCR analysis, which revealed a 518-bp hph-specific fragment with the primer pair F3/R3 (S3A and S4A Figs, S1 Table). As expected, the hph fragment was not found from the wild type (WT) control (S4A Fig). Similarly, a 981-bp FoSlt2-specific fragment was detected in WT but not from the ΔFoSlt2 mutants with the primer pair ZJ-F1/ZJ-R1 (S3A and S4A Figs, S1 Table). A 586-bp zeocin-specific fragment was detected in complemented strain ΔFoSlt2-c but not from the WT with primer pair Zeo-F/Zeo-R (S3A and S4B Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F1/ZJ-R1 and 3, 5, 7 and 9 with primer pair F3/R3. For the FoMkk2 gene, three FoMkk2 deletion mutants were also identified by PCR analysis. No fragment from WT was detected and a 518-bp hph-specific fragment from mutant ΔFoMkk2 was detected with primer pair F3/R3 (S3B and S4C Figs, S1 Table). A 1010-bp FoMkk2-specific fragment from WT was detected and no fragment from mutant ΔFoMkk2 was detected with primer pair ZJ-F3/ZJ-R3 (S3B and S4C Figs, S1 Table). No fragment from WT was detected and a 586-bp zeocin-specific fragment from complemented strain ΔFoMkk2-c was detected with primer pair Zeo-F/Zeo-R (S3B and S4D Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F3/ZJ-R3 and 3, 5, 7 and 9 with primer pair F3/R3. For the FoBck1 gene, three FoBck1 deletion mutants were also identified by PCR analysis. No fragment from WT was detected and a 518-bp hph-specific fragment from mutant ΔFoBck1 was detected with primer pair F3/R3 (S3C and S4E Figs, S1 Table). A 988-bp FoBck1-specific fragment from WT was detected and no fragment from mutant ΔFoBck1 was detected with primer pair ZJ-F5/ZJ-R5 (S3C and S4E Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F5/ZJ-R5 and 3, 5, 7 and 9 with primer pair F3/R3. Marker: DL5000.
(TIF)
(A). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoSlt2 using a 981-bp FoSlt2 fragment amplified with primer pair ZJ-F1/ZJ-R1 as a probe (S3A Fig), an expected 2.2-kb fragment from WT was detected and no fragment from mutant ΔFoSlt2 was detected. (B). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoMkk2 using a 1010-bp FoMkk2 fragment amplified with primer pair ZJ-F3/ZJ-R3 as a probe (S3B Fig), an expected 3.5-kb fragment from WT was detected and no fragment from mutant ΔFoMkk2 was detected. (C). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoBck1 using a 988-bp FoBck1 fragment amplified with primer pair ZJ-F5/ZJ-R5 as a probe (S3C Fig), an expected 3.3-kb fragment from WT was detected and no fragment from mutant ΔFoBck1 was detected. (D). Southern blot analysis of the NcoI-digested genomic DNA from WT, mutants ΔFoSlt2 and complemented strain ΔFoSlt2-c using a 940-bp FoSlt2 upstream fragment amplified with primer pair ZJ-F2/ZJ-R2 as a probe (S3A Fig), expected 3.3-kb and 2.5-kb fragments were detected from WT and mutant ΔFoSlt2, and 2.5-kb and 4.2-kb fragments were detected from complemented strain ΔFoSlt2-c, respectively. (E). Southern blot analysis of the HindIII and BamHI-digested genomic DNA from WT, mutants ΔFoMkk2 and complemented strain ΔFoMkk2-c using a 930-bp FoMkk2 upstream fragment amplified with primer pair ZJ-F4/ZJ-R4 as a probe (S3B Fig), expected 2.3-kb and 3.4-kb fragments were detected from WT and ΔFoMkk2, and 1.9-kb and 3.4-kb fragments were detected from complemented strain ΔFoMkk2-c, respectively. (F). Southern blot analysis of the ScaI-digested genomic DNA from WT and mutants ΔFoBck1 using a 899-bp FoBck1 upstream fragment amplified with primer pair ZJ-F6/ZJ-R6 as a probe (S3C Fig), an expected 6.8-kb and 5.9-kb fragment was detected from WT and ΔFoBck1, respectively.
(TIF)
Gene expression levels are represented relative to WT using quantitative real-time PCR. Error bars indicate the standard error from three replicates. Quantitative real-time PCR analysis was performed with RNA samples of the indicated strains, FoSlt2 gene using primer pair RT-F1/RT-R1 (S3A Fig, S1 Table), FoMkk2 gene using primer pair RT-F2/RT-R2 (S3B Fig, S1 Table), and FoBck1 gene using primer pair RT-F3/ RT-R3 (S3C Fig, S1 Table). The expression levels of FoSlt2, FoMkk2 and FoBck1 genes were not detected in corresponding mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, while the complemented strains ΔFoSlt2-c and ΔFoMkk2-c were completely restored to WT levels, respectively.
(TIF)
(DOC)
Acknowledgments
We thank Dr. Lynda Ciuffetti (Oregon State University, USA) for providing the vector pCT74 and Dr. Lianghui Ji (Temasek Life Sciences Laboratory, Singapore) for providing the vector pZGR1. We are grateful to Mr. Tinashe J. Runyanga for proof reading of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by Council of Agriculture, China. (No. CAR-32-05) and National Natural Science Foundation of China (31200112). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ploetz RC (2006) Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp cubense . Phytopathology 96: 653–656. 10.1094/PHYTO-96-0653 [DOI] [PubMed] [Google Scholar]
- 2. Li MH, Xie XL, Lin XF, Shi JX, Ding ZJ, Ling JF, et al. (2014) Functional characterization of the gene FoOCH1 encoding a putative α-1,6-mannosyltransferase in Fusarium oxysporum f. sp. cubense . Fungal Genet Biol 65: 1–13. 10.1016/j.fgb.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 3. Qi XZ, Guo LJ, Yang LY, Huang JS (2013) Foatf1, a bZIP transcription factor of Fusarium oxysporum f. sp cubense, is involved in pathogenesis by regulating the oxidative stress responses of Cavendish banana (Musa spp.). Physiol Mol Plant P 84: 76–85. [Google Scholar]
- 4. Zhao X, Kim Y, Park G, Xu JR (2005) A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea . Plant Cell 17: 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rispail N, Soanes DM, Ant C, Czajkowski R, Grunler A, Huguet R, et al. (2009) Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet Biol 46: 287–298. 10.1016/j.fgb.2009.01.002 [DOI] [PubMed] [Google Scholar]
- 6. Zeng FY, Gong XY, Hamid MI, Fu YP, Xie JT, Cheng JS, et al. (2012) A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet Biol 49: 347–357. 10.1016/j.fgb.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 7. Jain R, Valiante V, Remme N, Docimo T, Heinekamp T, Hertweck C, et al. (2011) The MAP kinase MpkA controls cell wall integrity, oxidative stress response, gliotoxin production and iron adaptation in Aspergillus fumigatus . Mol Microbiol 82: 39–53. 10.1111/j.1365-2958.2011.07778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valiante V, Jain R, Heinekamp T, Brakhage AA (2009) The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus . Fungal Genet Biol 46: 909–918. 10.1016/j.fgb.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 9. Zhao X, Mehrabi R, Xu JR (2007) Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot Cell 6: 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hou ZM, Xue CY, Peng YL, Katan T, Kistler HC, Xu JR. (2002) A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol Plant Microbe Interact 15: 1119–1127. [DOI] [PubMed] [Google Scholar]
- 11. Yago JI, Lin CH, Chung KR (2011) The SLT2 mitogen-activated protein kinase-mediated signalling pathway governs conidiation, morphogenesis, fungal virulence and production of toxin and melanin in the tangerine pathotype of Alternaria alternata . Mol Plant Pathol 12: 653–665. 10.1111/j.1364-3703.2010.00701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fourie G, Steenkamp ET, Ploetz RC, Gordon TR, Viljoen A (2011) Current status of the taxonomic position of Fusarium oxysporum formae specialis cubense within the Fusarium oxysporum complex. Infect Genet Evol 11: 533–542. 10.1016/j.meegid.2011.01.012 [DOI] [PubMed] [Google Scholar]
- 13. Di Pietro A, Garcia-MacEira FI, Meglecz E, Roncero MI (2001) A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol Microbiol 39: 1140–1152. [PubMed] [Google Scholar]
- 14. O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dong X, Ling N, Wang M, Shen QR, Guo SW (2012) Fusaric acid is a crucial factor in the disturbance of leaf water imbalance in Fusarium-infected banana plants. Plant Physiol Bioch 60: 171–179. 10.1016/j.plaphy.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 16. Schoffelmeer EA, Klis FM, Sietsma JH, Cornelissen BJ (1999) The cell wall of Fusarium oxysporum . Fungal Genet Biol 27: 275–282. [DOI] [PubMed] [Google Scholar]
- 17. Odenbach D, Thines E, Anke H, Foster AJ (2009) The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Mol Plant Pathol 10: 81–94. 10.1111/j.1364-3703.2008.00515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Brien JA, Daudi A, Butt VS, Bolwell GP (2012) Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236: 765–779. 10.1007/s00425-012-1696-9 [DOI] [PubMed] [Google Scholar]
- 19. Eisendle M, Oberegger H, Zadra I, Haas H (2003) The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding L-ornithine N-5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol Microbiol 49: 359–375. [DOI] [PubMed] [Google Scholar]
- 20. Logrieco A, Moretti A, Castella G, Kostecki M, Golinski P, Ritieni A, et al. (1998) Beauvericin production by Fusarium species. Appl Environ Microbiol 64: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Berges MS, Hera C, Sulyok M, Schafer K, Capilla J, Guarro J, et al. (2013) The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Mol Microbiol 87: 49–65. 10.1111/mmi.12082 [DOI] [PubMed] [Google Scholar]
- 22. Niehaus EM, von Bargen KW, Espino JJ, Pfannmuller A, Humpf HU, Tudzynski B. (2014) Characterization of the fusaric acid gene cluster in Fusarium fujikuroi . Appl Microbiol Biotechnol 98: 1749–1762. 10.1007/s00253-013-5453-1 [DOI] [PubMed] [Google Scholar]
- 23. Brown DW, Butchko RA, Busman M, Proctor RH (2012) Identification of gene clusters associated with fusaric acid, fusarin, and perithecial pigment production in Fusarium verticillioides . Fungal Genet Biol 49: 521–532. 10.1016/j.fgb.2012.05.010 [DOI] [PubMed] [Google Scholar]
- 24. Xu JR, Staiger CJ, Hamer JE (1998) Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci U S A 95: 12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rui O, Hahn M (2007) The Slt2-type MAP kinase Bmp3 of Botrytis cinerea is required for normal saprotrophic growth, conidiation, plant surface sensing and host tissue colonization. Mol Plant Pathol 8: 173–184. 10.1111/j.1364-3703.2007.00383.x [DOI] [PubMed] [Google Scholar]
- 26. Dirr F, Echtenacher B, Heesemann J, Hoffmann P, Ebel F, Wagener J. (2010) AfMkk2 is required for cell wall integrity signaling, adhesion, and full virulence of the human pathogen Aspergillus fumigatus . Int J Med Microbiol 300: 496–502. 10.1016/j.ijmm.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 27. Jeon J, Goh J, Yoo S, Chi MH, Choi J, Rho HS, et al. (2008) A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae . Mol Plant Microbe Interact 21: 525–534. 10.1094/MPMI-21-5-0525 [DOI] [PubMed] [Google Scholar]
- 28. Navarro-Garcia F, Alonso-Monge R, Rico H, Pla J, Sentandreu R, Nombela C. (1998) A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans . Microbiology 144 (Pt 2): 411–424. [DOI] [PubMed] [Google Scholar]
- 29. Muller S, Baldin C, Groth M, Guthke R, Kniemeyer O, Brakhage AA, et al. (2012) Comparison of transcriptome technologies in the pathogenic fungus Aspergillus fumigatus reveals novel insights into the genome and MpkA dependent gene expression. BMC Genomics 13: 519 10.1186/1471-2164-13-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valiante V, Heinekamp T, Jain R, Hartl A, Brakhage AA (2008) The mitogen-activated protein kinase MpkA of Aspergillus fumigatus regulates cell wall signaling and oxidative stress response. Fungal Genet Biol 45: 618–627. [DOI] [PubMed] [Google Scholar]
- 31. Lopez-Berges MS, Capilla J, Turra D, Schafferer L, Matthijs S, Jochl C, et al. (2012) HapX-Mediated Iron Homeostasis Is Essential for Rhizosphere Competence and Virulence of the Soilborne Pathogen Fusarium oxysporum . Plant Cell 24: 3805–3822. 10.1105/tpc.112.098624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schrettl M, Bignell E, Kragl C, Sabiha Y, Loss O, Eisendle M, et al. (2007) Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog 3: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schrettl M, Beckmann N, Varga J, Heinekamp T, Jacobsen ID, Jochl C, et al. (2010) HapX-Mediated Adaption to Iron Starvation Is Crucial for Virulence of Aspergillus fumigatus . PLoS Pathog 6: e1001124 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oide S, Moeder W, Krasnoff S, Gibson D, Haas H, Yoshioka K, et al. (2006) NPS6, encoding a nonribosomal peptide synthetase involved in siderophore-mediated iron metabolism, is a conserved virulence determinant of plant pathogenic ascomycetes. Plant Cell 18: 2836–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN, et al. (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigates virulence. J Exp Med 200: 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li C, Zuo C, Deng G, Kuang R, Yang Q, Hu C, et al. (2013) Contamination of bananas with beauvericin and fusaric acid produced by Fusarium oxysporum f. sp. cubense . PLoS One 8: e70226 10.1371/journal.pone.0070226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Desjardins AE, Proctor RH (2007) Molecular biology of Fusarium mycotoxins. Int J Food Microbiol 119: 47–50. [DOI] [PubMed] [Google Scholar]
- 38. Calvo AM, Wilson RA, Bok JW, Keller NP (2002) Relationship between secondary metabolism and fungal development. Microbiol Mol Biol R 66: 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hooykaas PJJ, Roobol C, Schilperoort RA. (1979) Regulation of the transfer of Ti plasmids of Agrobacterium tumefaciens . J Gen Microbiol 110: 99–109. [Google Scholar]
- 40. Li S, Myung K, Guse D, Donkin B, Proctor RH, Grayburn WS, et al. (2006) FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides . Mol Microbiol 62: 1418–1432. [DOI] [PubMed] [Google Scholar]
- 41. Lorang JM, Tuori RP, Martinez JP, Sawyer TL, Redman RS, Rollins JA, et al. (2001) Green fluorescent protein is lighting up fungal biology. Appl Environ Microbiol 67: 1987–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leng Y, Zhong S (2012) Sfp-type 4'-phosphopantetheinyl transferase is required for lysine synthesis, tolerance to oxidative stress and virulence in the plant pathogenic fungus Cochliobolus sativus . Mol Plant Pathol 13: 375–387. 10.1111/j.1364-3703.2011.00756.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Di Pietro A, Roncero MI (1998) Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum . Mol Plant Microbe Interact 11: 91–98. [DOI] [PubMed] [Google Scholar]
- 44. Fujioka T, Mizutani O, Furukawa K, Sato N, Yoshimi A, Yamagata Y, et al. (2007) MpkA-Dependent and-independent cell wall integrity signaling in Aspergillus nidulans . Eukaryot Cell 6: 1497–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Selvaggini S, Munro CA, Paschoud S, Sanglard D, Gow NA (2004) Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae . Microbiology 150: 921–928. [DOI] [PubMed] [Google Scholar]
- 46. Kapteyn JC, Hoyer LL, Hecht JE, Muller WH, Andel A, Verkleij AJ, et al. (2000) The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol 35: 601–611. [DOI] [PubMed] [Google Scholar]
- 47. Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23: 403–405. [DOI] [PubMed] [Google Scholar]
- 48. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
The GenBank accession numbers are Magnaporthe grisea Mps1 (AF020316), Aspergillus fumigatus MpkA (XM_746366, AFUA_4G13720), Colletotrichum lagenarium Maf1 (AY064246), Fusarium graminearum Mgv1 (AF492766), Aspergillus nidulans MpkA (U59214), Fusarium fujikuroi IMI 58289 MAP kinase (CCT68358), Fusarium verticillioides 7600 CMGC/MAPK/ERK1 protein kinase (EWG40764), Fusarium proliferatum putative MAP kinase (ABD67163), Fusarium oxysporum Fo5176 hypothetical protein FOXB_06615 (EGU82812), Aspergillus fumigatus Mkk2 (XM_745237, AFUA_1G05800), Neurospora crassa OR74A MAP kinase kinase (EAA28074), Fusarium oxysporum Fo5176 hypothetical protein FOXB_03604 (EGU85756), Fusarium oxysporum f. sp. melonis 26406 STE/STE7/MKK protein kinase (EXK41584), Fusarium fujikuroi IMI 58289 probable MAP kinase kinase (CCT66161), Fusarium verticillioides 7600 STE/STE7/MKK protein kinase (EWG44090), Magnaporthe oryzae 70–15 STE/STE7 protein kinase (XP_003717079), Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE7/MKK protein kinase (EXM04167), Magnaporthe grisea Mck1 (XP_368361), Aspergillus fumigatus Bck1 (XM_749418; AFUA_3G11080), Coniothyrium minitans Bck1 (JF951364), Fusarium oxysporum f. sp. lycopersici MN25 STE/STE11/BCK1 protein kinase (EWZ92886), Fusarium verticillioides 7600 STE/STE11/BCK1 protein kinase (EWG43584), Fusarium graminearum PH-1 hypothetical protein FGSG_06326 (ESU12405), Fusarium fujikuroi IMI 58289 bck1-like MAPKKK (CCT67356), Fusarium oxysporum f. sp. cubense tropical race 4 54006 STE/STE11/BCK1 protein kinase (EXM06456). All protein sequences were aligned using Clustal × 2.0. Aligned sequences were analyzed by Poisson model method in MEGA 5.2. Bootstrap values were calculated from 1000 bootstrap replicates.
(TIF)
(A) Strategic map of gene replacement and complementation construct, and restriction map of the FoSlt2 genomic region. (B) Strategic map of gene replacement and complementation construct, and restriction map of the FoMkk2 genomic region. (C) Strategic map of gene replacement and restriction map of the FoBck1 genomic region. The relative positions of the primers (short arrows) used for amplification of the linear DNA fragment employed for gene replacement, identification, quantitative real-time PCR and Southern blot analysis are indicated. H, HindIII; N, NcoI; B, BamHI; S, ScaI.
(TIF)
For the FoSlt2 gene, three FoSlt2 deletion mutants were identified by PCR analysis, which revealed a 518-bp hph-specific fragment with the primer pair F3/R3 (S3A and S4A Figs, S1 Table). As expected, the hph fragment was not found from the wild type (WT) control (S4A Fig). Similarly, a 981-bp FoSlt2-specific fragment was detected in WT but not from the ΔFoSlt2 mutants with the primer pair ZJ-F1/ZJ-R1 (S3A and S4A Figs, S1 Table). A 586-bp zeocin-specific fragment was detected in complemented strain ΔFoSlt2-c but not from the WT with primer pair Zeo-F/Zeo-R (S3A and S4B Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F1/ZJ-R1 and 3, 5, 7 and 9 with primer pair F3/R3. For the FoMkk2 gene, three FoMkk2 deletion mutants were also identified by PCR analysis. No fragment from WT was detected and a 518-bp hph-specific fragment from mutant ΔFoMkk2 was detected with primer pair F3/R3 (S3B and S4C Figs, S1 Table). A 1010-bp FoMkk2-specific fragment from WT was detected and no fragment from mutant ΔFoMkk2 was detected with primer pair ZJ-F3/ZJ-R3 (S3B and S4C Figs, S1 Table). No fragment from WT was detected and a 586-bp zeocin-specific fragment from complemented strain ΔFoMkk2-c was detected with primer pair Zeo-F/Zeo-R (S3B and S4D Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F3/ZJ-R3 and 3, 5, 7 and 9 with primer pair F3/R3. For the FoBck1 gene, three FoBck1 deletion mutants were also identified by PCR analysis. No fragment from WT was detected and a 518-bp hph-specific fragment from mutant ΔFoBck1 was detected with primer pair F3/R3 (S3C and S4E Figs, S1 Table). A 988-bp FoBck1-specific fragment from WT was detected and no fragment from mutant ΔFoBck1 was detected with primer pair ZJ-F5/ZJ-R5 (S3C and S4E Figs, S1 Table). Lanes: 2, 4, 6 and 8 with primer pair ZJ-F5/ZJ-R5 and 3, 5, 7 and 9 with primer pair F3/R3. Marker: DL5000.
(TIF)
(A). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoSlt2 using a 981-bp FoSlt2 fragment amplified with primer pair ZJ-F1/ZJ-R1 as a probe (S3A Fig), an expected 2.2-kb fragment from WT was detected and no fragment from mutant ΔFoSlt2 was detected. (B). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoMkk2 using a 1010-bp FoMkk2 fragment amplified with primer pair ZJ-F3/ZJ-R3 as a probe (S3B Fig), an expected 3.5-kb fragment from WT was detected and no fragment from mutant ΔFoMkk2 was detected. (C). Southern blot analysis of the HindIII-digested genomic DNA from WT and mutants ΔFoBck1 using a 988-bp FoBck1 fragment amplified with primer pair ZJ-F5/ZJ-R5 as a probe (S3C Fig), an expected 3.3-kb fragment from WT was detected and no fragment from mutant ΔFoBck1 was detected. (D). Southern blot analysis of the NcoI-digested genomic DNA from WT, mutants ΔFoSlt2 and complemented strain ΔFoSlt2-c using a 940-bp FoSlt2 upstream fragment amplified with primer pair ZJ-F2/ZJ-R2 as a probe (S3A Fig), expected 3.3-kb and 2.5-kb fragments were detected from WT and mutant ΔFoSlt2, and 2.5-kb and 4.2-kb fragments were detected from complemented strain ΔFoSlt2-c, respectively. (E). Southern blot analysis of the HindIII and BamHI-digested genomic DNA from WT, mutants ΔFoMkk2 and complemented strain ΔFoMkk2-c using a 930-bp FoMkk2 upstream fragment amplified with primer pair ZJ-F4/ZJ-R4 as a probe (S3B Fig), expected 2.3-kb and 3.4-kb fragments were detected from WT and ΔFoMkk2, and 1.9-kb and 3.4-kb fragments were detected from complemented strain ΔFoMkk2-c, respectively. (F). Southern blot analysis of the ScaI-digested genomic DNA from WT and mutants ΔFoBck1 using a 899-bp FoBck1 upstream fragment amplified with primer pair ZJ-F6/ZJ-R6 as a probe (S3C Fig), an expected 6.8-kb and 5.9-kb fragment was detected from WT and ΔFoBck1, respectively.
(TIF)
Gene expression levels are represented relative to WT using quantitative real-time PCR. Error bars indicate the standard error from three replicates. Quantitative real-time PCR analysis was performed with RNA samples of the indicated strains, FoSlt2 gene using primer pair RT-F1/RT-R1 (S3A Fig, S1 Table), FoMkk2 gene using primer pair RT-F2/RT-R2 (S3B Fig, S1 Table), and FoBck1 gene using primer pair RT-F3/ RT-R3 (S3C Fig, S1 Table). The expression levels of FoSlt2, FoMkk2 and FoBck1 genes were not detected in corresponding mutants ΔFoSlt2, ΔFoMkk2 and ΔFoBck1, while the complemented strains ΔFoSlt2-c and ΔFoMkk2-c were completely restored to WT levels, respectively.
(TIF)
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.