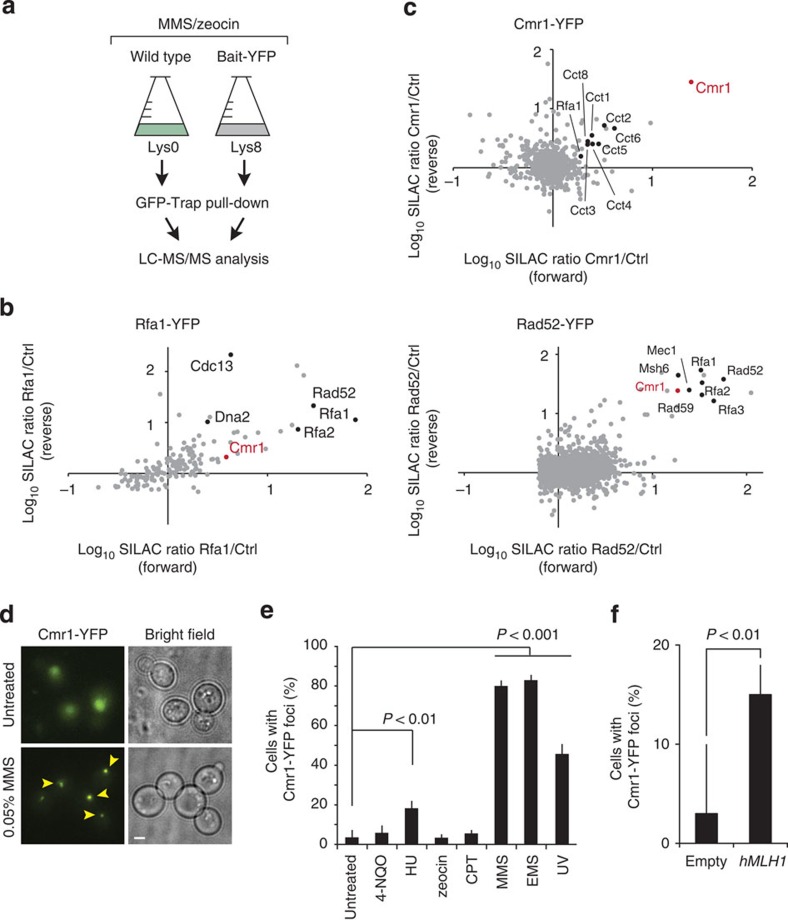
Figure 1. Cmr1 associates with RPA-bound chromatin.
(a) Representation of the workflow used for the SILAC-based identification of protein complexes associated with DNA repair factors. Yeast strains expressing YFP-tagged (IG54-11D and IG46-1B) or untagged (IG45-8A) proteins were cultured in SILAC media and harvested in log phase after treatment with DNA-damaging agents. Protein complexes from SILAC lysates were affinity purified separately with GFP-Trap. Proteins were trypsin proteolysed and peptides were identified by liquid chromatography (LC)–MS/MS. (b) Identification of Rfa1-YFP and Rad52-YFP interacting proteins. The plots show log(10) SILAC ratios from GFP-tagged bait versus control from forward and reverse (SILAC label swap) experiments. Dots indicate identified proteins. Cmr1 is highlighted in red and some of the known interactors for Rfa1 and Rad52 are indicated in black in the respective plots. (c) Identification of Cmr1 interacting proteins. Cmr1-YFP (IG71-2B) was used as bait for the pull down; Cmr1 is highlighted in red; CCT-chaperonin complex subunits and Rfa1 are indicated in black. (d) Cmr1 relocalization into foci. Representative images of untreated and MMS-treated cells are shown. Arrowheads indicate selected foci. Scale bar, 2 μm. (e) Quantification of Cmr1 foci. Cmr1-YFP localization was examined by fluorescence microscopy in IG66. Cells were grown to exponential phase and imaged after treatment with zeocin (200 μg ml−1), MMS (0.05%), CPT (5 μg ml−1), 4-NQO (0.2 μg ml−1), HU (200 mM), EMS (0.5%) for 2 h, or 1 h after ultraviolet irradiation (25 J m−2). Error bars represent 95% confidence intervals. Two to 3 replicates of 100–200 cells were analysed for each condition. (f) hMLH1 expression causes accumulation of Cmr1 foci. Cells expressing Cmr1-YFP (IG66) were transformed with pEH333 for ectopic expression of hMLH1 or with an empty vector (pEH334). Error bars represent 95% confidence intervals. Two to 3 replicates of 100–200 cells were analysed for each strain.