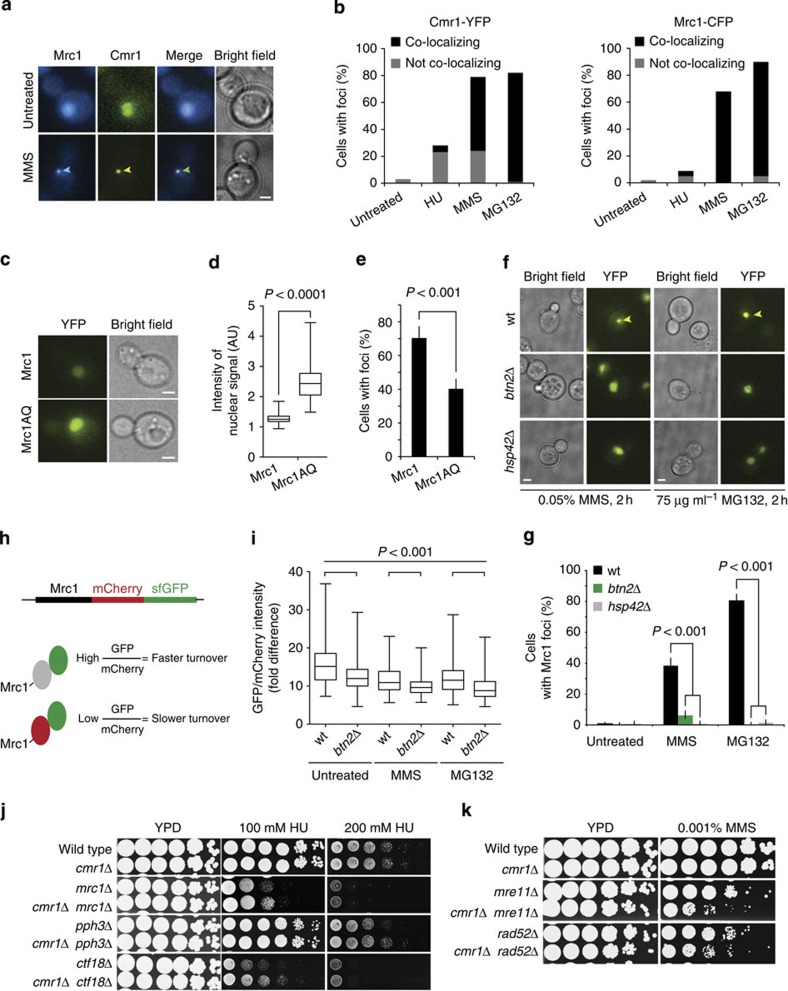
Figure 5. Cmr1 is involved in the response to replication stress.
(a) Mrc1 and Cmr1 foci co-localize during replication stress. Co-localization between Cmr1-YFP and Mrc1-CFP was assessed in untreated cells (IG160-4A) and after treatment with MMS for 2 h. Representative images are shown. Scale bar, 2 μm. Arrowhead indicates INQ focus. (b) Quantification of co-localization of Mrc1 and Cmr1 in response to HU, MMS and MG132 (n>200). (c) Checkpoint-defective Mrc1AQ protein accumulates in the nucleus. Mrc1-YFP (IG147) and Mrc1AQ-YFP (IG315) were imaged. Scale bars, 2 μm. (d) Quantification of Mrc1AQ protein levels. Images from c were quantified (n>150). The box plot displays fluorescence intensities in arbitrary units (AU), where the line across the box identifies the median sample value, the ends of the box are the 25th and 75th percentiles, and whiskers represent minimum and maximum values. (e) Checkpoint-defective Mrc1AQ exhibits reduced recruitment to INQ. Percentage of cells with Mrc1-YFP or Mrc1AQ-YFP foci was quantified after treatment with 0.05% MMS for 2 h. Error bars represent 95% confidence intervals (n>150). (f) Mrc1 focus formation requires BTN2 and HSP42. Mrc1-YFP localization was assessed in wild-type (IG147), btn2Δ (CC1–3B) and hsp42Δ (CC2–6B) cells. Representative images of Mrc1 localization are shown. Scale bars, 2 μm. (g) Quantification of Mrc1 foci in hsp42Δ and btn2Δ mutants shown in f. Error bars represent 95% confidence intervals. Two replicates, n>250. (h) Schematic representation of Mrc1 fusion to a fluorescent timer. The ratio between the sfGFP (fast maturing) and mCherry (slow maturing) fluorescence intensities was calculated as a measure of Mrc1 protein turnover. (i) Quantification of Mrc1 protein turnover. The fold difference between the GFP and mCherry nuclear fluorescence was measured in wild-type (CC98) and btn2Δ (CC102-9C) strains. Box plot were displayed as in d. Two replicates, n>100. (j) CMR1 deletion suppresses the DNA damage sensitivity of checkpoint mutants. Tenfold serial dilutions were plated on YPD or YPD containing the indicated drug. Strains were ML8–9A (wt), DP1 (cmr1Δ), IG156-7D (mrc1Δ), IG156-6C (mrc1Δ cmr1Δ), IG257-9C (pph3Δ), IG257-2C (pph3Δ cmr1Δ), IG177-9C (ctf18Δ) and IG177-8C (ctf18Δ cmr1Δ). (k) cmr1Δ is synthetic sick with homologous recombination mutants. Tenfold serial dilutions were plated on YPD or YPD containing 0.001% MMS. Strains were ML8-9A (wt), DP1 (cmr1Δ), IG164-1D (mre11Δ), IG164-2B (mre11Δ cmr1Δ), IG162-1D (rad52Δ) and IG162-2D (rad52Δ cmr1Δ).