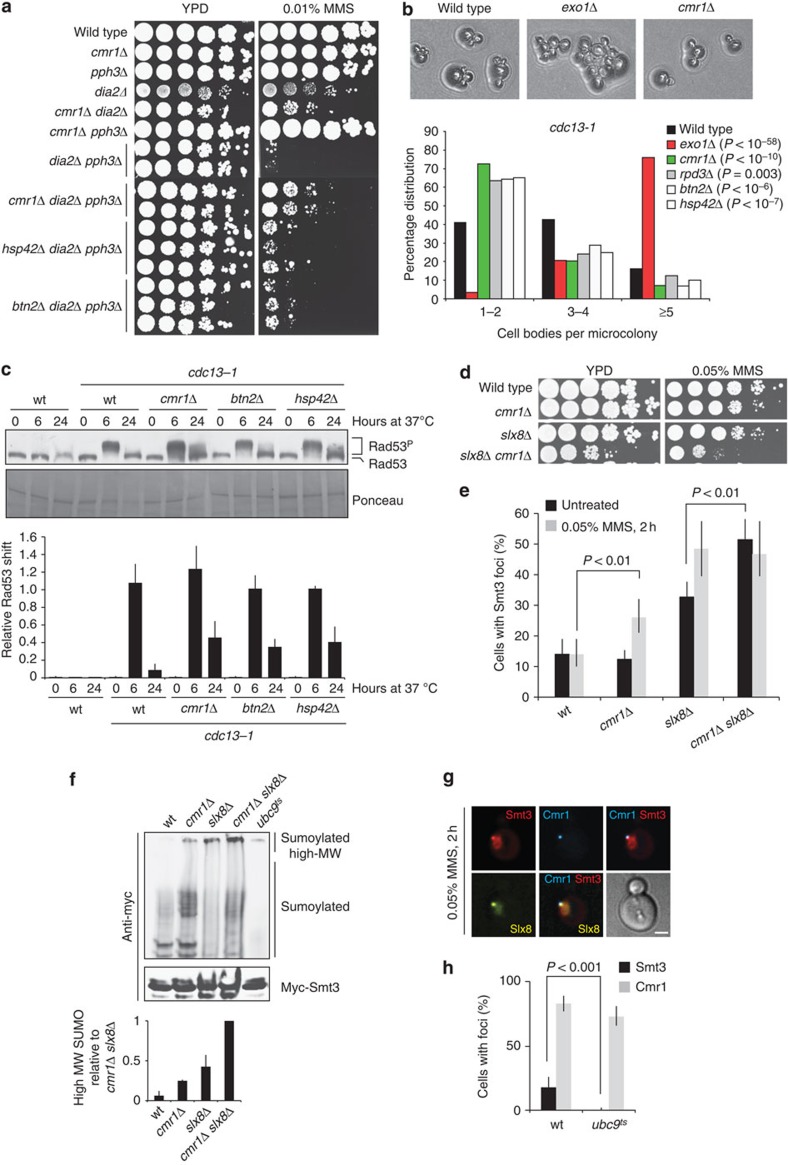
Figure 6. Cmr1 promotes adaptation to the DNA damage checkpoint.
(a) Cmr1 genetic interaction with checkpoint deactivation pathways is functionally associated with INQ. Wild-type (ML8–9A), cmr1Δ (DP1), dia2Δ (CC4–19D), dia2Δ cmr1Δ (CC4-3B), pph3Δ (IG257-9C), cmr1Δ pph3Δ (IG257-2C), dia2Δ pph3Δ (IG296-2C), cmr1Δ dia2Δ pph3Δ (IG296-49B), hsp42Δ pph3Δ dia2Δ (IG322-12A) and btn2Δ pph3Δ dia2Δ (IG323-19D) strains were plated. (b) Mutants of INQ are checkpoint adaptation defective. Strains with a conditional cdc13-1 mutation and otherwise wild type (ML815-8A), exo1Δ (DLY1296), cmr1Δ (ML808-12D), rpd3Δ (ML807-2D), btn2Δ (ML821-4C) or hsp42Δ (ML822-4C) were examined. (c) Mutants of INQ are defective in Rad53 dephosphorylation during adaptation. cdc13-1 mutants were grown at 25 °C before being shifted to the restrictive temperature of 37 °C. Samples were harvested at 0, 6 and 24 h after temperature shift. Rad53 phosphorylation was detected by immunoblotting and the relative shift (Rad53P/(Rad53+Rad53P)) for each of the samples was quantified in the lower panel. Error bars reflect the s.d. of two independent experiments. (d) Negative genetic interaction between cmr1Δ and slx8Δ. Wild-type (ML8–9A), cmr1Δ (DP1), slx8Δ (NEB290-1B) and cmr1Δ slx8Δ (IG256-5D) strains were plated. (e) Cmr1 and Slx8 promote turnover of SUMO foci. Wild-type, cmr1Δ, slx8Δ and cmr1Δ slx8Δ cells (same strains as in d) ectopically expressing yEmRFP-Smt3 (pML133) were imaged. Percentage of cells with Smt3 foci was quantified. Error bars represent 95% confidence intervals (n>100). (f) Sumoylated and polysumoylated proteins accumulate in the cmr1Δ mutant. Cells from the experiment in d were transformed with a plasmid expressing 3myc-Smt3 (pRS313-3myc-Smt3), harvested in log phase and immunoprecipitation using anti-myc-coupled dynabeads was performed on whole-cell extracts. The temperature-sensitive ubc9-1 mutant (IG246-2C) grown at 37 °C was used as a negative control. Bands corresponding to high-molecular weight (high-MW) SUMO conjugates were quantified from two independent experiments using ImageJ. Error bars indicate s.d. (g) Co-localization of Slx8 and SUMO at INQ. Cells expressing Cmr1-CFP, Slx8-YFP and yEmRFP-Smt3 (IG302-1D transformed with pML133) were imaged. Representative image of co-localization between Smt3 and Slx8 at INQ (Cmr1) is shown. Scale bar, 2 μm. (h) INQ contains sumoylated proteins. Cmr1 and Smt3 localization was monitored in wild type (IG66) and ubc9ts mutant (IG246-2C) after incubation at 37 °C for 2 h in the presence of 0.05% MMS. Error bars represent 95% confidence intervals (n>100).