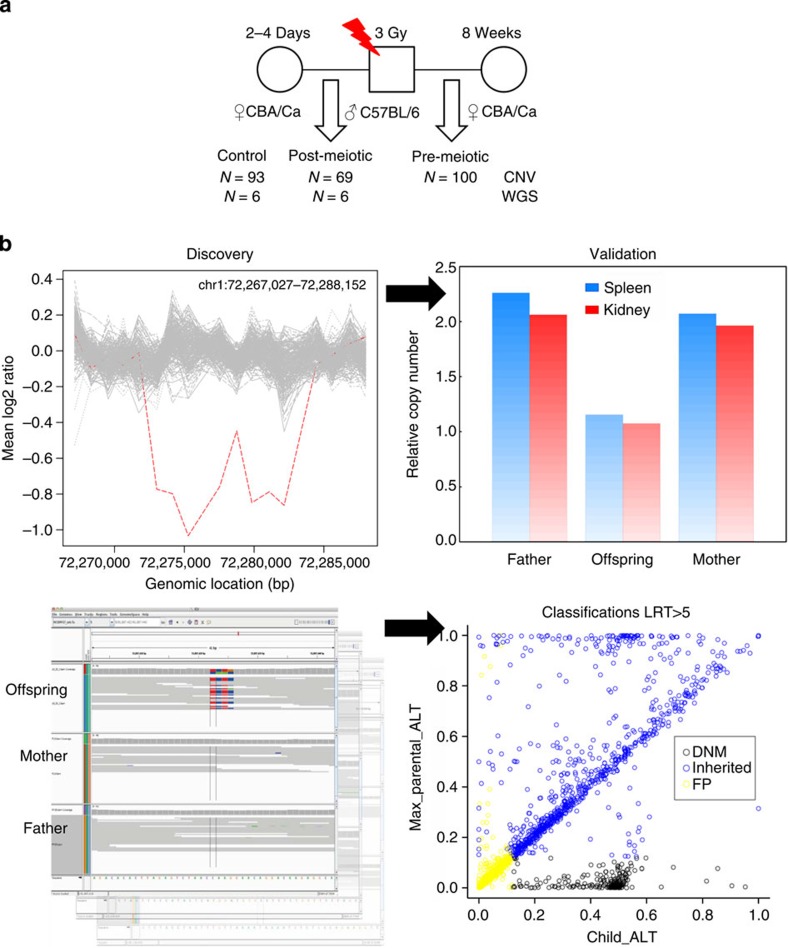
Figure 1. Experimental design and discovery of de novo mutations.
(a) Design of mutational study. The number of offspring analysed by comparative genome hybridization (CNV) and whole-genome sequencing (WGS) is shown. (b) Discovery and validation of de novo mutations. The panel depicts a representative CNV profile with a 9.1-kb deletion (left) validated by qPCR (right). The bottom panel displays a sequence alignment and discovery of a multisite variant in one offspring (left), and validation of all putative de novo SNVs and indels in all offspring (right), where variants are classified as true de novo variants, inherited, or false positive categories based on read count proportions of the alternative allele.