Figure 1.
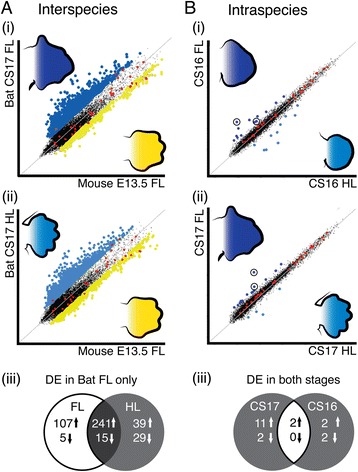
A microarray comparison of bat forelimb and hindlimb autopod transcript abundance with the mouse forelimb. (A) Two direct analyses between the array signal from (i) CS17 bat (blue) forelimb (FL) and the mouse (yellow) FL and (ii) CS17 bat hindlimb (HL) and the mouse FL revealed many genes that were significantly differentially expressed (DE). (iii) Commonly DE genes were removed from these lists to identify 112 genes that were uniquely DE between the CS17 FL and the mouse FL. (B) Two indirect analyses comparing the bat FL and HL at stages (i) CS16 and (ii) CS17 revealed few differences between the array signal of these limb types. Hoxd11 and Meis2 (circled) were identified as highly DE in these comparisons. (iii) Commonly DE genes were found by comparing gene lists using a fold change (FC) filter of either FC > 2 (interspecies) or FC > 1.5 (intraspecies) and a cut-off value of q < 0.01.
