Figure 3.
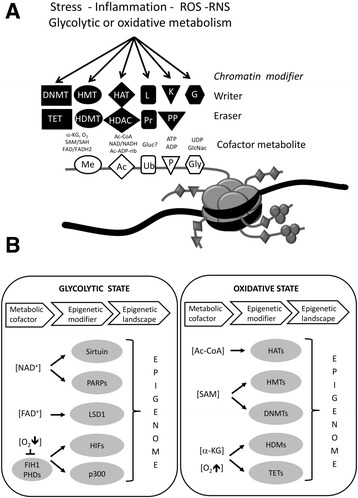
Activity of chromatin modifying writer-eraser enzymes depends on available concentrations of cofactor metabolites and environmental signals. (A) Schematic representation of a nucleosome with extruding histone tails with residues that can be modified by various chromatin writer (i.e., DNA methyltransferase (DNMT), histone methyltransferase (HMT), histone acetylase (HAT), ubiquitin ligase (L), kinase (K), glycosylase (G)) or chromatin eraser enzymes (i.e., DNA hydroxymethylase (TET), demethylase (HDMT), deacetylase (HDAC), proteasome (Pr), phosphatase (PP)), resulting in dynamic histone methylation (Me), acetylation (Ac), ubiquitination (Ub), phosphorylation (P), and glycosylation (Gly). These histone modifications have been associated with changes in chromatin organization, gene activation, silencing, and several other nuclear functions (adapted from [338]). (B) Hypothetical model of a glycolytic-oxidative metabolic switch and its possible influence on epigenetic modifiers and the epigenetic landscape (adapted from [339]).
