Figure 2.
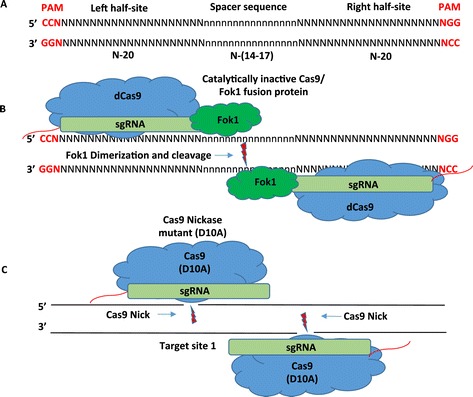
Dual CRISPR-Cas technologies that increase nuclease specificity. (A, B) Dimeric CRISPR RNA-guided Fok1 nuclease target sequences consist of two 20 nt half-sites flanked by a protospacer adjacent motif (PAM) sequence in the form of 5′-NGG that are separated by a 14–17 nt spacer sequence. Each half-site is bound by a Cas-9/Fok1 fusion protein. Once bound, the Fok1 domains of two different Cas-9/Fok1 fusion proteins dimerize and introduce a double-stranded break in the spacer sequence. (C) Dual RNA-guided CRISPR-Cas9 nickase system. In this system, two sgRNAs are expressed that each guide a mutant version of Cas9 (Cas9-D10A) (that only nicks one strand of the DNA rather than making a double-stranded cut) to two different sequences that flank the target region. The two Cas9 nickases bind to opposite strands of the DNA nicking both DNA strands flanking the target region. This introduces a site-specific double-stranded break that is then repaired by NHEJ.
