Abstract
Burkholderia pseudomallei is an intracellular pathogen and the causative agent of melioidosis, a severe disease of humans and animals. One of the virulence factors critical for early stages of infection is the Burkholderia secretion apparatus (Bsa) Type 3 Secretion System (T3SS), a molecular syringe that injects bacterial proteins, called effectors, into eukaryotic cells where they subvert cellular functions to the benefit of the bacteria. Although the Bsa T3SS itself is known to be important for invasion, intracellular replication, and virulence, only a few genuine effector proteins have been identified and the complete repertoire of proteins secreted by the system has not yet been fully characterized. We constructed a mutant lacking bsaP, a homolog of the T3SS “gatekeeper” family of proteins that exert control over the timing and magnitude of effector protein secretion. Mutants lacking BsaP, or the T3SS translocon protein BipD, were observed to hypersecrete the known Bsa effector protein BopE, providing evidence of their role in post-translational control of the Bsa T3SS and representing key reagents for the identification of its secreted substrates. Isobaric Tags for Relative and Absolute Quantification (iTRAQ), a gel-free quantitative proteomics technique, was used to compare the secreted protein profiles of the Bsa T3SS hypersecreting mutants of B. pseudomallei with the isogenic parent strain and a bsaZ mutant incapable of effector protein secretion. Our study provides one of the most comprehensive core secretomes of B. pseudomallei described to date and identified 26 putative Bsa-dependent secreted proteins that may be considered candidate effectors. Two of these proteins, BprD and BapA, were validated as novel effector proteins secreted by the Bsa T3SS of B. pseudomallei.
Burkholderia pseudomallei is a Gram-negative environmental saprophyte found in free-standing water and soil (1). The causative agent of melioidosis, B. pseudomallei causes severe disease in humans and animals with a range of clinical manifestations including severe pneumonia and septic shock, acute or chronic suppurative infections, genitourinary disease, and asymptomatic carriage (2). An emerging infectious disease, melioidosis is found primarily in South East Asia and Northern Australia, yet there is a growing body of evidence of a more widespread distribution in subtropical regions of Asia, the Americas, and Africa (3). Melioidosis is an important cause of community-acquired septicaemia in North-East Thailand, where it can cause mortality rates of up to 40% (4). Because of the lack of an effective vaccine, intrinsic antibiotic resistance, and the high mortality rates, there is a need to better understand the molecular mechanisms of pathogenesis to inform the design of treatments and vaccines (5).
B. pseudomallei is a facultative intracellular pathogen capable of invading both phagocytic and nonphagocytic cells (6). Once inside the cell, the bacteria can rapidly escape the endosome and polymerize host cell actin at one pole causing actin-based motility, a process dependent on the bacterial BimA protein (7). B. pseudomallei also causes host cell fusion creating multinucleated giant cells (MNGCs) that facilitates cell to cell spread while evading immune detection (8). The formation of MNGCs by B. pseudomallei is dependent on secretion of the protein VgrG5 by the virulence-associated type VI secretion system-5 (9–12).
One important virulence factor of B. pseudomallei that has also been shown to be important for infection in many other Gram-negative bacteria including pathogenic Escherichia coli, Salmonella, Shigella, and Yersinia, is the Type III Secretion System (T3SS)1 (13–16). T3SSs act as a molecular syringe projecting outwards from the surface of the bacteria allowing for the transport of bacterial proteins into target eukaryotic cells (17). These bacterial proteins, termed effectors, subvert host cell processes to the advantage of the bacteria and rely on a second class of T3SS-secreted proteins (translocators) for delivery (18). B. pseudomallei contains three T3SSs, of which T3SS-1 and T3SS-2 share homology with T3SSs found in plant pathogens (19, 20). T3SS-3, also known as the Burkholderia Secretion Apparatus (Bsa) T3SS, is similar to the Inv-Spa (Salmonella pathogenicity island (SPI)-1) and Mxi-Spa T3SSs of the animal pathogens Salmonella spp. and Shigella flexneri respectively (21). The Bsa T3SS has been shown to be important in invasion, endosome escape and net intracellular replication in cultured cells and in virulence in murine and Syrian hamster models of melioidosis (21–24).
As the Bsa system influences pathogenesis, one may infer that its effectors also influence the outcome of infection. However, only three effectors, BopE, BopC, and CHBP, have so far been proven to be secreted by B. pseudomallei in a Bsa T3SS-dependent manner (25–27). BopE is a guanine nucleotide exchange factor that is involved in invasion of nonphagocytic cells (26). BopC has been shown to be important for invasion of A549 epithelial cells and endosome escape, and intracellular survival in J774A.1 macrophage-like cells (25, 28). CHBP is a homolog of E. coli Cif, a cyclomodulin that disrupts the eukaryotic cell cycle (29), for which evidence of Bsa-dependent translocation into host cells was recently presented (27). A fourth candidate effector, BopA, prevents LC3-associated host autophagy of intracellular bacteria and has been shown to be secreted in a T3SS-dependent manner in a surrogate enteropathogenic E. coli (EPEC) host (30, 31). Compared with the large number of effectors secreted by T3SSs in other Gram-negative pathogens, it is likely that there are other effectors of the Bsa T3SS yet to be characterized (32). Bsa-secreted proteins are known to be potent B- and T-cell antigens in humans (33–36) and subunit vaccines based on needle components or effectors are protective against other pathogens that deploy T3SSs (e.g. Shigella IpaB and IpaD, Yersinia V antigen, Salmonella SseI) (37–41).
One of the problems facing T3SS effector protein discovery is the tight regulation of the T3SS under laboratory conditions with different T3SSs responding to very different environmental cues (42–47). Although some genes involved in transcriptional control of the Bsa T3SS were recently identified (48), little is known about the post-translational regulation of the system and what environmental signals may activate it. Indeed, a recent proteomics study of B. pseudomallei failed to identify any of the known Bsa T3SS effectors in bacterial culture supernatants (49). To override the tight control of the T3SS, one solution employed in the study of other Gram-negative bacteria (for example EPEC and Citrobacter rodentium) was to generate mutations of T3SS components that dysregulate the system, thereby creating a hypersecretion phenotype. The bacterial supernatants were then analyzed using gel-free quantitative proteomics techniques to identify a range of novel effector proteins (50, 51).
In Shigella, deletion of IpaD, the needle tip protein, creates a “leaky” phenotype, in which higher levels of both translocators and effectors are seen in the supernatant (46, 52). The homolog of IpaD in B. pseudomallei is BipD (53) (supplemental Fig. S1). In a previous study, we have shown a 10276 bipD::pDM4 mutant secreted higher levels of the effector protein BopE into the bacterial culture supernatant by Western blot analysis, but it is unknown what effect bipD disruption may have on the secretion of other effectors and translocators (26).
Another group of T3SS proteins that have been shown in many systems to be involved in the control of effector and translocator secretion are the so called “gatekeeper” family of proteins, InvE/MxiC/SepL/YopN-TyeA of Salmonella/Shigella/E. coli/Yersinia, respectively (17). Investigation of these proteins has given insight into the temporal regulation of their respective T3SSs. Although deletion of all of these proteins causes an increase in levels of secreted effectors there are also differences between them. Deletion of InvE and SepL cause a reduction in secreted levels of translocators, deletion of MxiC has no effect on their levels, and deletion of YopN increases levels of secreted translocators (54–60). The closest homolog to this family of proteins in B. pseudomallei is the uncharacterized protein BsaP (BPSS1544) (supplemental Fig. S1).
Here, a 10276 ΔbsaP deletion mutant was generated and its effect on secreted levels of the known effector BopE was assessed. To define the total and Bsa-dependent repertoire of secreted proteins, Isobaric Tags for Relative and Absolute Quantification (iTRAQ), a gel-free quantitative proteomics technique, was used to quantify protein levels in the bacterial culture supernatants of B. pseudomallei mutant strains 10276 bipD::pDM4 and 10276 ΔbsaP. In order to identify candidate Bsa effector proteins, the secreted protein profiles from these hypersecreting mutants were then compared with the isogenic parent strain (10276 WT) and a T3SS structural-null mutant (10276 bsaZ::pDM4), which has been shown to be unable to secrete BipD and BopE (21). BsaZ is a homolog of YscU in Yersinia, which forms a key component of the inner membrane ring and is required for a functional T3SS (61) (supplemental Fig. S1). From this analysis we provide the first evidence of the involvement of both BipD and BsaP in control of known effector/translocator secretion in the Bsa T3SS. We also present a comprehensive general secretome for B. pseudomallei. Furthermore, we have identified 26 candidate effector proteins, including all known Bsa-secreted effectors, of which we show two proteins are novel substrates for secretion by the Bsa T3SS.
EXPERIMENTAL PROCEDURES
Strains, Media, Plasmids, and Oligonucleotides
Bacterial strains and plasmids are listed in supplemental Table S1. The insertion mutants 10276 bipD::pDM4 and 10276 bsaZ::pDM4 were produced by integration of the pDM4 suicide vector into the coding sequence of the target gene (21). Primers, purchased from Sigma Aldrich (Gillingham, UK), are listed in supplemental Table S2. Bacteria were routinely cultured at 37 °C on LB agar (Miller) or LB broth (Lennox). Antibiotic selection was performed at the following concentrations unless otherwise stated: ampicillin (Amp) 100 μg/ml, kanamycin (Kan) 50 μg/ml, chloramphenicol (Cm) 34 μg/ml, and tetracycline (Tet) 15 μg/ml. IPTG was used at a final concentration of 0.5 mm where appropriate. Unless otherwise stated all chemicals were purchased from Sigma Aldrich (Gillingham, UK).
B. pseudomallei Mutagenesis
The 10276 ΔbsaP in-frame deletion mutant was made using allelic exchange and the sacB containing positive-selection suicide vector pDM4 essentially as previously described (62). DNA fragments upstream and downstream of bsaP (BPSS1544) were amplified by PCR from B. pseudomallei 10276 genomic DNA using primer pair's bsaP-del-1/bsaP-del-2 and bsaP-del-3/bsaP-del-4. The resulting DNA fragments were combined using PCR-ligation-PCR (63). A-tailing was performed to add an adenosine to the blunt-ended PCR product, which was then ligated into pGEM-T resulting in the plasmid pGEM-T-ΔbsaP. The insert was sequenced and a SpeI and BglII fragment containing the 10276 ΔbsaP fragment ligated into similarly digested pDM4 resulting in pDM4-ΔbsaP. To propagate pDM4-ΔbsaP, the plasmid was transformed into TOP 10 PIR1 cells (Life Technologies, Paisley, UK) before transfer into E. coli S17–1 λpir conjugal donor with selection for Cm resistance.
pDM4-ΔbsaP was introduced into B. pseudomallei strain 10276 by conjugation with E. coli S1–17 λpir and plated onto LA plates containing Cm (50 μg/ml) and Kan (50 μg/ml) to select for B. pseudomallei with pDM4-ΔbsaP integrated into the chromosome by homologous recombination (21). Colonies were patched onto LA plates containing Cm (50 μg/ml) and Kan (50 μg/ml) to confirm resistance. Overnight cultures of the Cmr colonies were cultured without selection to allow for a second round of homologous recombination to take place either restoring the full length bsaP gene or resulting in bsaP deletion. To select for double recombinants lacking pDM4-ΔbsaP the overnight cultures were plated onto 15% sucrose plates lacking NaCl and cultured at 30 °C for 3 days. Chromosomal deletion of bsaP was confirmed in sucrose resistant/Cms colonies by colony PCR and the resulting strain designated 10276 ΔbsaP, where the bsaP gene has been replaced by an in-frame start-stop (atg-taa) cassette.
Complementation of the ΔbsaP Mutant
For complementation of the ΔbsaP mutant the 10276 bsaP sequence was amplified by PCR using the forward primer bsaP-exp-F containing a ClaI restriction site and a ribosome-binding site and the reverse primer bsaP-exp-R containing a HindIII restriction site. The PCR fragment was cloned into pBHR1 (MoBiTec, 2B Scientific Ltd., Oxford, UK) to create pBHR1-bsaP. The plasmid was purified and transformed into B. pseudomallei 10276 ΔbsaP using the fast sucrose method of electroporation (64), creating strain 10276 ΔbsaP-pbsaP.
Cloning of Candidate Effectors to Express c-Myc Epitope-Tagged Fusion Proteins
The coding region for candidate effectors based on the B. pseudomallei K96243 genome sequence were amplified from B. pseudomallei 10276 genomic DNA by PCR with a 5′ forward primer containing a ribosome-binding site and a 3′ reverse primer encoding an in-frame c-Myc tag followed by a stop codon. Each primer also contained a restriction site suitable for directional insertion into the vector pME6032. Plasmids were transformed into E. coli and the inserts sequenced. Purified plasmids were then transformed into B. pseudomallei 10276 (WT), 10276 bsaZ::pDM4, 10276 bipD::pDM4, and 10276 ΔbsaP.
Preparation of Bacterial Culture Supernatants
Preparation of B. pseudomallei supernatants was performed by first centrifuging overnight cultures at 3220 RCF for 10 min at 4 °C followed by disposal of the supernatant and resuspension of the bacteria in fresh LB to an OD600 of 1.0. Of the bacterial suspension, 1 ml was then added to 10 ml of LB containing the appropriate antibiotics and IPTG if needed. The culture was incubated for 6 h shaking at 200 RPM at 37 °C. To determine the initial and final bacterial counts, a sample was removed from the culture before and after the 6 h incubation for serial dilution and plating on LA plates. After 6 h, the sample was centrifuged at 3220 RCF for 10 min at 4 °C. The culture supernatant was collected and centrifuged a second time to ensure no intact bacteria remained. The supernatant was then passed through a 0.2 μm low protein binding cellulose acetate membrane filter (Millipore, Watford, UK) and sterility tested.
Protein Precipitation
Proteins were precipitated using a Pyrogallol red-molybdate methanol (PRMM) solution (0.5 mm pyrogallol red (Alfa Aesar, Heysham, UK), 0.16 mm sodium molybdate, 1.0 mm sodium oxalate, 50 mm succinic acid, 20% MeOH, pH 2.0) as previously described (65). In brief, equal volumes of bacterial culture supernatant and PRMM solution were thoroughly mixed and the pH was adjusted to 2.8 ± 0.1 with HCl. The samples were then precipitated overnight at 4 °C. The resulting precipitate was centrifuged at 3220 RCF, 4 °C for 1 h and the supernatant was discarded. The pellet was washed twice with ice-cold acetone and then allowed to air dry. The protein pellet was then solubilized in 50 mm ammonium bicarbonate and quantified using a Direct Detect Infrared Spectrometer (Millipore, Watford, UK).
SDS-PAGE and Immunoblotting
Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Equal volumes of protein samples and Laemmli buffer were mixed and heated to 90 °C for 5 min. Samples were separated on Any kDa™ Mini-PROTEAN® TGX gels (Bio-Rad, Hemel Hempstead, UK) at 150V for 45 min in Tris-Glycine running buffer. Proteins were visualized by Coomassie staining. Western blots were used to identify particular proteins of interest. Proteins were transferred onto a nitrocellulose membrane using the Trans-Blot Turbo system (Bio-Rad, Hemel Hempstead, UK) at 2.5 A for 3 min. α-BopE (26), α-BipD (22), or α-cMyc (Insight Biotechnology, Wembley, UK) rabbit polyclonal antibodies were used at 1 μg/ml. For detection by infrared fluorescence, goat- α -rabbit IgG (H+L) (DyLight™ 800 Conjugate) was used at a 1:10,000 dilution (Cell Signaling Technology, Hitchin, UK). Blots were visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences, Cambridge, UK) according to the manufacturer's instructions. Images were captured and analyzed using Image Studio Lite (LI-COR Biosciences, Cambridge, UK).
iTRAQ Experimental Design
Bacterial culture supernatants of B. pseudomallei 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4, and 10276 ΔbsaP were generated in three independent experiments. Equal amounts of protein from each replicate were combined creating a single pooled sample for each strain. These samples were enzymatically digested, peptides were labeled using iTRAQ reagents, and then pooled and submitted to LC MS/MS in three technical replicates as follows:
(1) Protein Digestion and Peptide Fractionation
Protein samples were prepared for MS analysis using a protocol adapted from (66). Following protein quantification, samples were reduced in 10 mm DTT and alkylated in 50 mm Iodoacetamide prior to heating in Laemmli buffer, and then separated by one-dimensional SDS-PAGE (4–12% Bis-Tris Novex mini-gel, Life Technologies, Paisley, UK) and visualized by colloidal Coomassie staining (Life Technologies). The entire protein gel lanes were excised and cut into 10 slices each. Every gel slice was subjected to in-gel digestion with trypsin overnight at 37 °C. The resulting tryptic peptides were extracted by formic acid (1%) and acetonitrile, lyophilized in a speedvac, and resuspended in 1% formic acid.
(2) Peptide Concentration
To determine the concentration of the peptides, the dried samples were resuspended in 100 mm TEAB 1 m ACN. To determine the volume of sample needed for an even protein load across all samples, 1 μl was injected onto an LTQ Velos Mass Spectrometer (Thermo Fisher Scientific, MA) and the TIC trace was used.
(3) iTRAQ Labeling
Samples were labeled according to the manufacturer's instructions using an iTRAQ Reagents Multiplex kit – four-plex (AB Sciex UK Ltd., Warrington, UK). To each vial of labeling reagent (labels- 114, 115, 116, and 117), 70 μl ethanol was added, after which they were vortexed and centrifuged. A single vial of label was added to a single vial of sample and mixed. The samples used with each label were: 10276 bsaZ::pDM4–114, 10276 WT-115, 10276 bipD::pDM4–116, and 10276 ΔbsaP-117. Samples were incubated with shaking at room temperature for 1 h, after which 100 μl of water was added and incubated for a further hour. The four vials of labels/samples were pooled into one vial and then dried under vacuum. Samples were desalted using a standard method.
(4) SCX Fractionation – Salt Cuts
To a Millipore (Watford, UK) C18 ziptip, 10 μl Poros HS resin (Life Technologies, Paisley, UK) was added and washed with 3 × 25 μl Buffer A (35% acetonitrile (ACN), 0.1% formic acid (FA)). Samples were resuspended in Buffer A and loaded onto the resin, washed with 3 × 25 μl Buffer A, and eluted with 2 × 25 μl of each of 11 salt cuts ranging from 5 mm to 800 mm NaCl. A final elution was performed using 2 × 25 μl 50% isopropanol, 0.4% ammonium hydroxide. Samples were centrifuged and speed vacuumed until dry. Samples were resuspended in 1% FA and pooled as six samples containing the salt cuts as follows: JS-1 (5 mm, 10 mm, 20 mm), JS-2 (50 mm), JS-3 (100 mm), JS-4 (150 mm), JS-5 (200 mm), and JS-6 (250 mm, 300 mm, 400 mm, 800 mm).
Mass Spectrometry
Peptides were separated using an Ultimate 3000 RSLC nanoflow LC system (Thermo Fisher Scientific, MA). With a constant flow of 5 μl/min, 15 μl of sample was loaded onto an Acclaim PepMap100 nanoViper C18 trap column (100 μm inner-diameter, 2 cm) (Thermo Fisher Scientific, MA). After trap enrichment, peptides were eluted onto an Acclaim PepMap RSLC nanoViper, C18 column (75 μm, 15 cm) (Thermo Fisher Scientific, MA) with a linear gradient of 2–45% solvent B (80% ACN with 0.08% FA) over 125 min with a constant flow of 300 nl/min. The HPLC system was coupled to a linear ion trap Orbitrap hybrid mass spectrometer (LTQ-Orbitrap Velos) (Thermo Fisher Scientific, MA) via a nanoelectrospray ion source (Thermo Fisher Scientific, MA). The spray voltage was set to 1.2kV and the temperature of the heated capillary was set to 250 °C. Full-scan MS survey spectra (m/z 335–1800) in profile mode were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 1,000,000 ions. The ten most intense peptide ions from the preview scan in the Orbitrap were fragmented sequentially by higher-energy collisional dissociation (HCD) (two-step (6%) normalized collision energy (NCE), 40%; and activation time, 0.1 ms) prior to analysis in the Orbitrap after accumulation of 50,000 ions. Maximal filling times were 500 ms for the full scans and 200 ms for the MS/MS scans. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged species were rejected. The lock mass option was enabled for survey scans to improve mass accuracy (67). Data were acquired using Xcalibur software (Thermo Fisher Scientific, MA).
Quantification and Bioinformatic Analysis
The raw mass spectrometric data files obtained for each experiment were collated into a single data set using Proteome Discoverer version 2.0 (Thermo Fisher Scientific, MA) and the Mascot search engine version 2.4 (www.matrixscience.com) using a translated B. pseudomallei K96243 genome as a reference for protein identification. Enzyme specificity was set to that of trypsin, allowing for cleavage N-terminal to proline residues and between aspartic acid and proline residues. Other parameters used were: 1) variable modifications, methionine oxidation, protein N-acetylation, gln → pyro-glu; 2) fixed modifications, cysteine carbamidomethylation; 3) database: B. pseudomallei K96243_20120601 (compiled 01/06/2012, containing 5728 proteins); 4) iTRAQ labels: standard iTRAQ 4-plex quant method with labels 114–117; 5) MS/MS tolerance: FTMS- 10ppm, FTMS/MS- 0.06Da; 6) minimum peptide length, 6; 7) maximum missed cleavages, 2; and 8) false discovery rate, 1%. Peptide ratios were calculated for samples using the 10276 bsaZ::pDM4–114 label as the denominator. Data are available via ProteomeXchange with identifier PXD001656.
Protein ratios were normalized using the overall median ratio for all quantified proteins in each sample based on the assumption that the abundance of the majority of secreted proteins was identical in each condition. Proteins were selected as potential T3SS effector protein candidates if they had at least two unique peptides used in quantification with a MASCOT peptide score ≥21 (which corresponds to the threshold value for a 95% confidence level, a 1 in 20 chance that the match is random), and a “high” iTRAQ ratio in at least one strain, which we defined as a value greater than 1.5 × IQR. IQR is defined as ; where n complete set of ratios over 10276 bsaZ::pDM4 for all quantified proteins in a given sample.
RESULTS
Construction and Characterization of Hypersecreting bsa Mutants
We hypothesized that BsaP may control the timing and magnitude of effector secretion based on homology to “gatekeeper” proteins from the T3SSs of other Gram-negative bacterial pathogens. BLAST analysis of available genomes indicated that BsaP shares 33% identity over 92% coverage (E-value 1e-41) with the Shigella MxiC “gatekeeper” protein and 34% identity over 88% coverage (E-value 4e-32) with the Salmonella InvE “gatekeeper” protein (supplemental Fig. S2). A nonpolar bsaP deletion mutant was therefore constructed in B. pseudomallei strain 10276 by double homologous recombination using a positive-selection suicide vector containing short regions of DNA flanking the bpss1544 gene (62). The chromosomal deletion was confirmed by sequencing across the bsaP adjoining regions (data not shown). Because bsaP is predicted to reside in the middle of an operon, with seven genes upstream and two downstream, the deletion of bsaP was complemented using a constitutive expression vector encoding bsaP to confirm any phenotypes were not caused by polar effects. Alongside the bsaP mutant we also analyzed 10276 bipD::pDM4 lacking the BipD needle tip protein that has been previously reported to secrete elevated levels of the effector protein BopE (26).
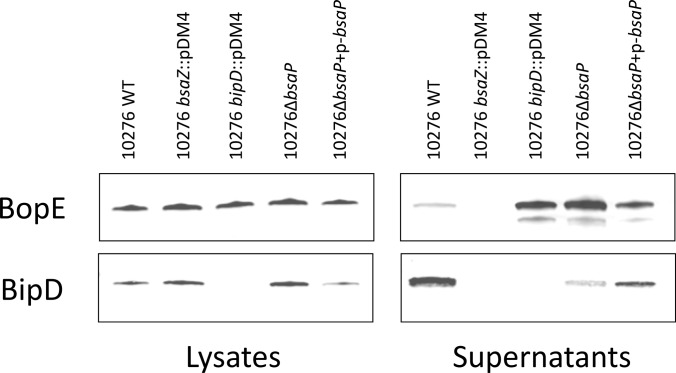
Bacteria-free culture supernatants were collected from B. pseudomallei 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4, 10276 ΔbsaP, and 10276 ΔbsaP (pbsaP) and analyzed by SDS-PAGE followed by Coomassie staining. The secreted protein profile of B. pseudomallei LB-culture supernatant is highly complex with few obvious differences between the strains (supplemental Fig. S3). The bacterial supernatants and whole cell lysates were also analyzed by Western blot using antibodies to detect the BopE effector protein and the BipD translocon protein (Fig. 1). The absence of both BopE and BipD in the supernatants of the 10276 bsaZ::pDM4 insertion mutant indicates that there was no detectable cell lysis at the time when the secretome was sampled. When compared with the 10276 WT strain, both 10276 bipD::pDM4 and 10276 ΔbsaP strains demonstrated increased secretion of BopE. Interestingly, the levels of BipD secreted into the supernatant was reduced in 10276 ΔbsaP compared with 10276 WT. This pattern of increased levels of the effector BopE and reduced secretion of the translocator BipD provides the first evidence of a role for BsaP as a “gatekeeper” protein, controlling a switch between the secretion of translocators and effectors. In the case of both BopE and BipD, the phenotype of 10276 ΔbsaP was partially restored by plasmid-mediated complementation, indicating the effects are unlikely to be caused by polar or secondary defects. Differences in the levels of BopE and BipD across the different strains sampled are not caused by differences in the number of bacteria at six hours subculture, because the growth rates of all strains were shown to be identical by viable count assessment (data not shown).
Fig. 1.
Western blot analysis of the expression and secretion of BopE and BipD proteins by wild-type and mutant B. pseudomallei strains. 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4, 10276 ΔbsaP, and 10276 ΔbsaP pbsaP were cultured for 6 h at 37 °C. Equal quantities of protein from the whole cell lysates or cell-free supernatants were separated by denaturing SDS-PAGE and proteins transferred to a nitrocellulose membrane for protein detection using polyclonal rabbit α-BopE or α-BipD (26) essentially as described under “Experimental Procedures.”
Characterization of the total B. pseudomallei Secretome under Standard Laboratory Conditions
Having identified two bsa mutant strains with dysregulated BopE secretion profiles in bacterial culture supernatants, we then sought to identify the total secreted protein profile from these two strains in comparison to the isogenic parent strain (10276) and a bsa-null mutant strain. In three independent experiments, culture supernatants were prepared from the four test strains at 6 h post subculture. The strains used were B. pseudomallei 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4, and 10276 ΔbsaP (supplemental Fig. S1). The samples were all assessed for a lack of cell lysis by immunoblotting for BopE. All 10276 bsaZ::pDM4 samples lacked BopE, indicating the samples were suitable for analysis (supplemental Fig. S4). After mixing equimolar amounts of each of the three biological replicates, the samples were subjected to iTRAQ labeling and MS/MS analysis as described under “Experimental Procedures.” iTRAQ allowed the simultaneous identification and relative quantification of peptide fragments in all four separate samples, resulting in the positive identification of a total of 475 proteins in all samples (listed in supplemental Table S3). The data was expressed as ratios of relative protein abundance either in the 10276 WT, 10276 bipD::pDM4, or 10276 ΔbsaP samples over the 10276 bsaZ::pDM4 sample as a denominator (i.e. 10276 WT/10276 bsaZ::pDM4, 10276 bipD::pDM4/10276 bsaZ::pDM4, and 10276 ΔbsaP/10276 bsaZ::pDM4). The ratios were normalized to the median for each strain and a density plot showed that the majority of protein ratios cluster around the median of one, indicating that the mutations made did not radically change the secretion profile of the bulk of proteins (supplemental Fig. S5). From this analysis we identified a core set of 426 proteins, out of the predicted total proteome of B. pseudomallei K96243 of ∼5900 proteins (68), that were present in all samples. These are listed in supplemental Table S4. Strikingly, the majority of these proteins (∼80%) are encoded by Chromosome 1.
Of the proteins identified in the core secretome, 54 are annotated with reference to the K92643 genome as hypothetical (uncharacterized) proteins, several more are annotated with metabolic functions (supplemental Table S4). The core secretome also consisted of several elongation factors (Ts, G, P, Tu) and heat shock/chaperone proteins (GroES, GroEL), many of which have previously been reported to be present on the surface of B. pseudomallei or part of the outer membrane proteome and recognized by the sera of recovering melioidosis patients (35, 69–71). In addition to the presence of a collagenase, chitobiase, bacterioferritin, and Sec secretory pathway constituents, the secretome also harbored several proteases and lipoproteins. Several phage components, Type I fimbriae, and flagellar subunits were also present, although there was a notable lack of any of the predicted T6SS structural or effector proteins, Type V family of autotransporters or the Type IV pilus protein PilA.
Prediction of the Bsa Effector Repertoire
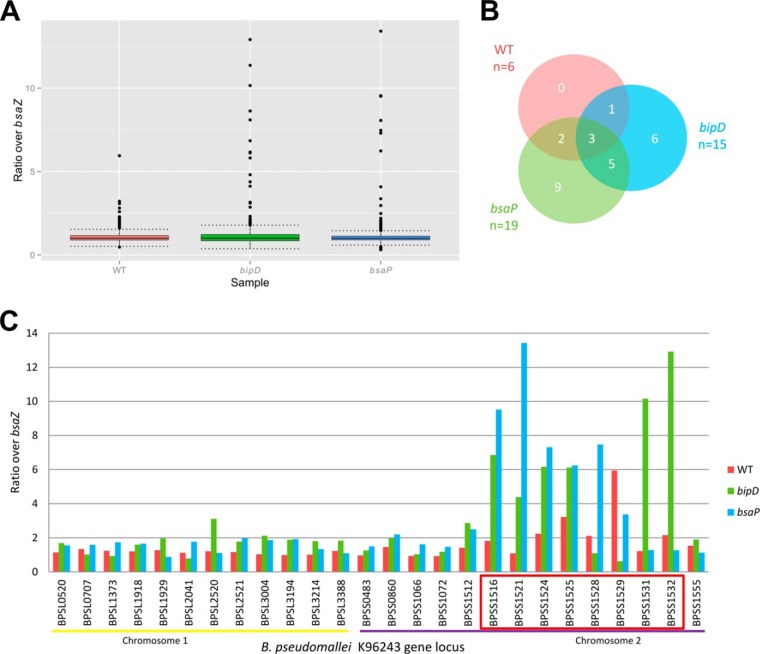
Putative T3SS effector proteins were identified in the data sets primarily on the calculated iTRAQ protein abundance ratios. The protein abundances obtained in each of the WT or mutant strain samples were each expressed as a ratio of the 10276 bsaZ::pDM4 sample and proteins with an iTRAQ ratio higher than the 1.5 × interquartile range (IQR), together with at least two unique peptides with a MASCOT peptide score ≥21, were taken to be T3SS effector protein candidates. The 1.5 × IQR values were calculated and the cut-offs set at 1.56 for the 10276 WT/10276 bsaZ::pDM4 data set, 1.79 for the 10276 bipD::pDM4/10276 bsaZ::pDM4 data set, and 1.46 for the 10276 ΔbsaP/10276 bsaZ::pDM4 data set, based on the calculation described under “Experimental Procedures.” When the data was plotted as a boxplot, the data showed a trend toward more proteins with larger ratios in the 10276 bipD::pDM4/10276 bsaZ::pDM4 and 10276 ΔbsaP/10276 bsaZ::pDM4 data sets when compared with the 10276 WT/10276 bsaZ::pDM4 data set (Fig. 2A). A total of 26 proteins met the selection criteria and can be considered putative effectors (Table I). Of the 26 proteins there were strain-specific subsets, with eight proteins with high ratios in two of the data sets and three proteins with high ratios in all three (Fig. 2B). The three proteins that displayed high ratios in all three data sets, BopC, BopA, and BopE, are all known B. pseudomallei Type III secreted (T3S) effector proteins or predicted to be T3S proteins based on their T3SS-dependent secretion in a surrogate host system (25, 26, 31) (Fig. 2B). Of the 26 proteins, 20 were only found in the hypersecreting mutant strain samples and would not have been identified had our study focused solely on the study of the secreted proteins of the 10276 WT strain (Fig. 2B). Plotting the iTRAQ ratios of the 26 putative effector proteins against their B. pseudomallei K96243 genetic locus number, it is clear that there is a concentration of eight proteins with especially high ratios encoded within the annotated bsa T3SS locus on Chromosome 2 compared with that of the rest of the genome (Table I, Fig. 2C).
Fig. 2.
Proteins with high iTRAQ protein abundance ratios are enriched in hypersecreting B. pseudomallei strains and encoded within the bsa T3SS locus. A, Boxplot generated by R software (www.r-project.org) showing the distribution and median values of the ratios of the protein abundances in the 10276 WT (WT), 10276 bipD::pDM4 (bipD), or 10276 ΔbsaP (bsaP) samples compared with the 10276 bsaZ::pDM4 (bsaZ) sample. The “cut-off” values (calculated using R software) are shown by the dotted lines. Points shown above the “cut-off” value are candidate T3SS effectors. B, A Venn diagram (constructed with Venny: www.bioinfogp.cnb.csic.es/tools/venny) depicting the number of strain-specific and shared candidate T3SS effector proteins identified in each of the strains 10276 WT (WT), 10276 bipD::pDM4 (bipD), and 10276 ΔbsaP (bsaP). C, Distribution of the candidate T3SS effector proteins plotted according to B. pseudomallei gene number across both Chromosomes and their ratios. Gene numbers refer to the B. pseudomallei K96243 reference strain. The bsa T3SS locus is indicated by the red box.
Table I. Candidate Bsa T3SS effector proteins identified by iTRAQ. The table lists proteins in either the 10276 WT, 10276 bipD::pDM4 or 10276 ΔbsaP culture supernatant samples that displayed protein abundance ratios (compared to the 10276 bsaZ::pDM4 mutant) higher than the calculated cut-off values for each comparison. The cut-off values were set at 1.56 for the 10276 WT/10276 bsaZ::pDM4 dataset, 1.79 for the 10276 bipD::pDM4/10276 bsaZ::pDM4 data set, and 1.46 for the 10276 ΔbsaP/10276 bsaZ::pDM4 data set, as described in “Experimental Procedures.” Additional criteria for protein identification included the presence of at least two unique peptides with a MASCOT peptide score of ≥21 corresponding to a 95% confidence level. The proteins are listed in order of gene number with genes from Chromosome one listed first. Those proteins fulfilling the criteria for selection as candidate T3SS effector proteins are highlighted in the gray shaded boxes. Protein score refers to the adjusted sum of scores of the individual peptide scores as calculated by MASCOT. Number of unique peptides refers to the number of significant peptides specific to a protein used in quantification.
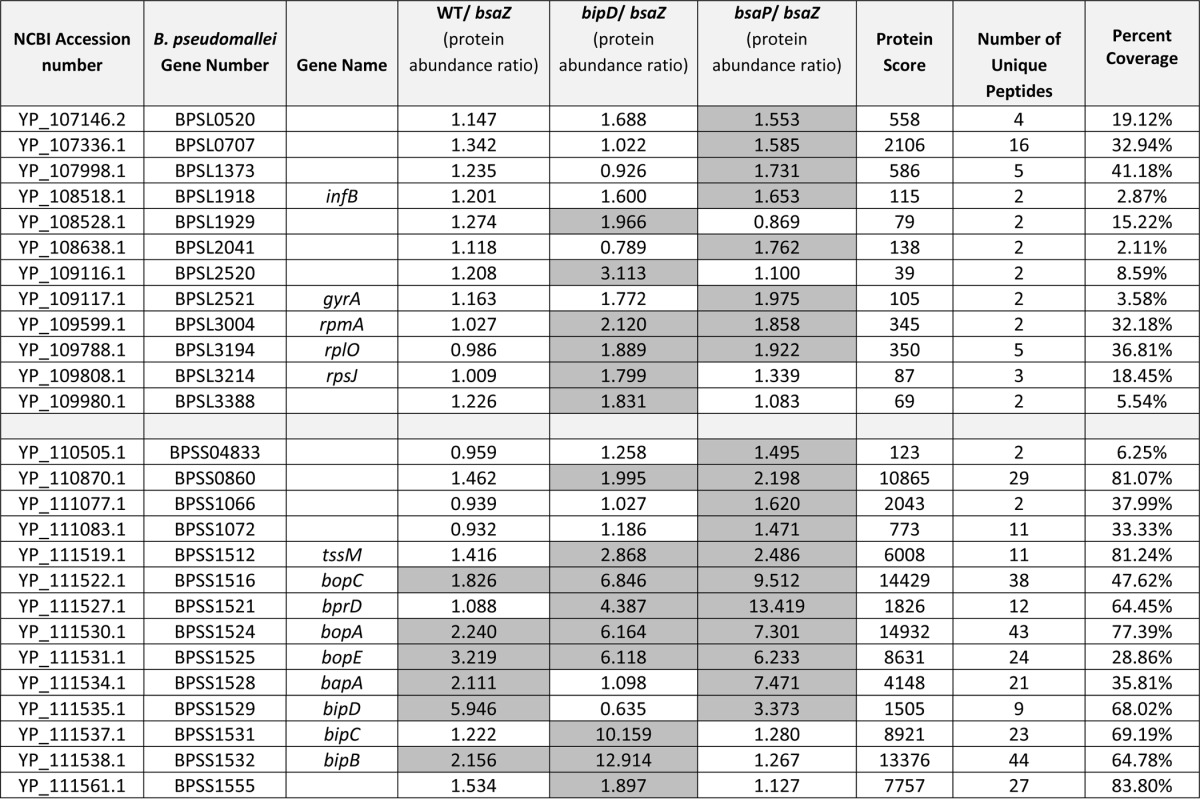
As expected we observed Bsa-dependent secretion of BopE in the 10276 WT strain that increased in 10726 bipD::pDM4 and 10276ΔbsaP strains, in full agreement with Western blot data (Fig. 1). BopC and BopA, the latter of which has only been proven to be secreted in a T3SS-dependent manner in EPEC, followed a similar pattern of secretion to BopE. Although both of the two putative translocator proteins, BipB and BipC, were secreted at higher levels in 10276 WT than 10276 bsaZ::pDM4, only BipB was secreted at levels above the cut-off ratio of 1.56. Similar to the effect of Shigella ipaD mutation, the mutation of bipD in B. pseudomallei 10276 caused an increase in secretion of both BipB and BipC into the supernatant compared with 10276 WT. Where 10276 bipD::pDM4 had increased ratios of the translocators, 10276ΔbsaP showed a decrease in ratios below that of 10276 WT in agreement with Western blot data (Fig. 1). This is further evidence that BsaP plays a similar role in B. pseudomallei to its homolog InvE in Salmonella as a “gatekeeper” protein. The only proven effector missing from this screen was CHBP, however this is not surprising as we have recently reported that the gene encoding CHBP, bpss1385, is absent from the genome of the B. pseudomallei 10276 strain used here (27).
Besides the needle tip protein BipD, both putative translocators BipB and BipC, BopA, BopC, and BopE, there are also two hypothetical proteins encoded within the bsa T3SS locus identified by the screen that have high iTRAQ ratios. The first, BapA (BPSS1528), is a predicted 880 amino acid protein with no conserved domains and homologs only in the closely related species B. mallei and B. thailandensis. Unlike the known effector proteins in the iTRAQ screen, although secretion of BapA appears to be Bsa-dependent in 10276 WT and the ratio increases in the 10276 ΔbsaP data set, it does not appear to be secreted abundantly by the 10276 bipD::pDM4 mutant. Interestingly, under the conditions used we did not detect BapB or BapC, which are encoded downstream of BapA in the same operon and predicted to be substrates of the T3SS (21).
The other candidate effector protein encoded in the bsa T3SS locus is BprD (BPSS1521), which is annotated as a hypothetical regulator (48). Predicted to be much smaller than BapA at 147aa, BprD is another protein with no known conserved domains and homologs that exist only within closely related Burkholderia species. The iTRAQ ratios suggest that it is not secreted at detectable levels by 10276 WT B. pseudomallei, but secreted by both 10276 bipD::pDM4 and 10276ΔbsaP, with the ratio of 13.4 in the 10276 ΔbsaP/10276 bsaZ::pDM4 data set being the highest of any protein described in this iTRAQ study.
Outside of the Bsa T3SS locus the remaining 18 proteins with high ratios are distributed across both B. pseudomallei chromosomes. One of the proteins encoded by a gene closely flanking the bsa locus is the virulence factor TssM (BPSS1512), which is known to be coregulated with the Bsa T3SS as well as secreted in a Type II-dependent manner (72).
Validation of iTRAQ Data by Analysis of the Secretion of Epitope-Tagged Candidate Effector Proteins
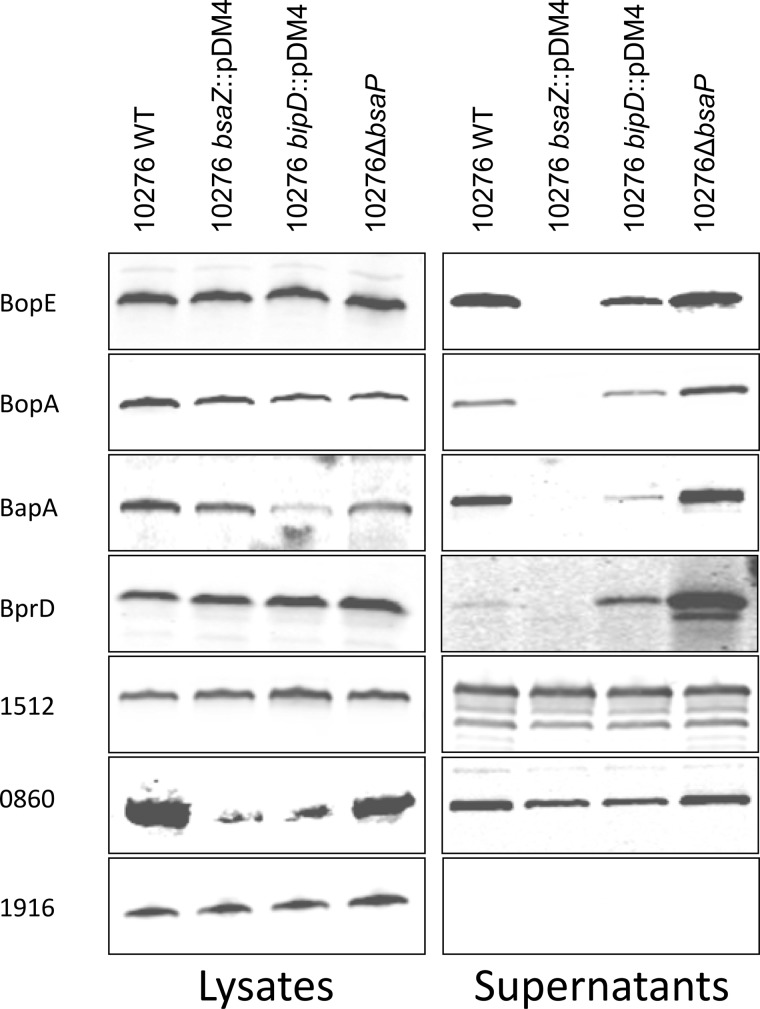
To confirm a number of targets are secreted in a Bsa-dependent manner, a number of candidates were selected and cloned into an inducible expression vector as in-frame C-terminal c-Myc tagged fusion proteins. The targets chosen were BprD, BapA, BPSS0860, BPSS1512, BopE, and BopA, as well as BPSS1916, a protein with a very high iTRAQ ratio in all strains that did not meet the initial selection criteria because of being quantified with only a single unique peptide. Targets were chosen to represent proteins with both high and low protein scores, high and low numbers of peptides, and to be encoded inside and outside of the bsa T3SS locus. The plasmids were transformed into strains 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4 and 10276ΔbsaP, and bacterial supernatants were collected and assessed by Western blot for the presence of the tagged proteins using a c-Myc tag-specific polyclonal antibody. Initial studies in which BopE was cloned as a c-Myc fusion in the widely used shuttle vector pBHR1 under a constitutive chloramphenicol resistance gene promoter indicated that BopE-c-Myc was secreted in a BsaZ-independent manner, possibly owing to the high level of expression (data not shown). We therefore elected to clone effector-c-Myc fusions under an IPTG-inducible promoter in pME6032 and select inducer levels at which strict BsaZ-dependent BopE-c-Myc secretion was observed (Fig. 3). BopA-c-Myc was absent in the supernatant of the 10276 bsaZ::pDM4 strain, but secreted by the parent strain and secreted at elevated levels by 10276ΔbsaP (Fig. 3), confirming for the first time in B. pseudomallei that it is a substrate of the Bsa T3SS, not just the locus of enterocyte effacement-encoded T3SS in EPEC (31). Furthermore, both of the putative effector proteins located within the bsa locus, BprD-c-Myc and BapA-c-Myc, were secreted in a BsaZ-dependent manner (Fig. 3), confirming that these are novel effectors of the Bsa apparatus.
Fig. 3.
Western blot analysis of the expression and secretion of putative B. pseudomallei Bsa T3SS effector proteins expressed as c-Myc tagged fusion proteins. The coding region of bopE, bopA, BPSS0860, BPSS1512, BPSS1916, bprD, and bapA were expressed with a c-terminal c-Myc tag in the IPTG-inducible expression vector pME6032 in B. pseudomallei strains 10276 WT, 10276 bsaZ::pDM4, 10276 bipD::pDM4 or 10276 ΔbsaP. Supernatant and whole cell fractions were collected after 6 h of incubation at 37 °C in the presence of 0.5 mm IPTG. Equal quantities of protein were separated by reducing SDS-PAGE, blotted onto nitrocellulose membranes and probed using polyclonal rabbit α-c-Myc antibody.
Interestingly, we were unable to validate Bsa-dependent secretion of proteins encoded outside of the Bsa T3SS locus. BPSS0860 and BPSS1512 appeared in the supernatants of all strains, including 10276 bsaZ::pDM4, indicating that although they are secreted, the Bsa system is not required for their secretion (Fig. 3). Secretion of the protein BPSS1916 could not be detected in any of the strains tested (Fig. 3). These examples highlight the need for independent validation of such data sets.
DISCUSSION
B. pseudomallei is an important cause of invasive clinical disease in South-East Asia and Northern Australia and its potential as a bioterrorism agent is cause for concern. One of the known virulence factors that is important for invasion, intracellular net replication and virulence in mice is the Bsa T3SS (21–24), but the repertoire of effector proteins secreted by the system has not yet been fully characterized. To establish a better understanding of the Bsa T3SS, we made mutations in key structural components shown to dysregulate other T3SSs. We show these mutations lead to hypersecretion of known effector proteins. Using these mutants we have performed a quantitative proteomic analysis of the total secretome of the Bsa T3SS system. This iTRAQ analysis identified all known effector proteins and translocators present in B. pseudomallei 10276, as well as identifying a number of novel putative effector proteins, two of which were independently validated. Also, examining the changes in the abundance of effectors and translocators secreted by the different mutants provides insight into the role the proteins likely play in the temporal regulation of the Bsa T3SS compared with other T3SSs of other Gram-negative bacteria.
The inability of previous proteomics studies to identify known Bsa T3SS effectors highlights the need for the use of more sensitive techniques such as iTRAQ in combination with genetic dysregulation. The B. pseudomallei secretome is very complex (supplemental Table S4, supplemental Fig. S3). This is not surprising when taking into account the large 7.2 Mb genome (68), which encodes ∼5900 proteins, of which over 55% are expressed in LB (73). This study produced one of the most comprehensive secretomes of B. pseudomallei to date, identifying a core secretome of 426 proteins secreted under standard laboratory conditions.
Although known effectors were absent from the secreted proteome of the bsaZ mutant by Western blotting (Fig. 1) and largely undetectable by iTRAQ (Table I) implying a lack of cell lysis, we detected a number of ribosomal proteins that are enriched in the supernatant when compared with the supernatant harvested from the 10276 bsaZ::pDM4 strain. This observation has been made in several other proteomic screens of T3SS effector repertoires (50, 74–76) and may be because of a combination of the high abundance of ribosomal proteins in the bacterial cell coupled with the sensitivity of iTRAQ.
Concurrent to this study, a proteome analysis investigating Type II secreted proteins of B. pseudomallei strain MSHR688 was published (72). From our screen, 27 of the 38 proteins identified as being substrates of Type II secretion in LB by Burtnick et al. (72) were also identified in our B. pseudomallei 10276 culture supernatants. Whether all 38 Type II secreted proteins identified in B. pseudomallei MSHR688 are encoded by B. pseudomallei 10276 is unknown because the genome sequence for B. pseudomallei 10726 is not yet available. Interestingly, three of the Type II secreted proteins were enriched in the supernatants of the hypersecreting mutants (BPSL0707, BPSS1512, BPSS1555) and two of the proteins are encoded by regions flanking the bsa T3SS locus, BPSS1512 and BPSS1555. The deubiquitinase BPSS1512 (TssM) has been shown to be secreted inside host cells where it suppresses the host innate immune response (77, 78). Although TssM is secreted independently of the Bsa T3SS, its expression appears to be coregulated with the Bsa T3SS and T6SS-5 (78, 79). This coregulation may explain the increased levels of TssM in the supernatants of the hypersecreting mutants in this study and again highlights the importance of validating candidate effector proteins.
The profile of secreted proteins encoded within the bsa T3SS locus provides a comprehensive picture of the secreted levels of the three known effector proteins. The profiles of both BopE and BopC in the iTRAQ screen correspond with previous studies in which they were shown to be secreted in a Bsa-dependent manner (25, 26). Although BopA has been shown to be Type 3 secreted in a surrogate host (31), we have confirmed its secretion to be dependent on the Bsa T3SS in B. pseudomallei and not one of the other T3SSs.
Two of the putative Bsa T3SS substrates encoded by the bsa T3SS locus (BapA and BprD) were validated as Bsa-dependent by Western blotting for a C-terminal c-Myc tag. Although the function of BapA is currently unknown, a bapA insertion mutation was not attenuated in a Syrian hamster model of acute melioidosis (23). BprD was previously annotated as a transcriptional regulator (48). In a previous study, when bprD was deleted along with bprB and bprC, there was no change in levels of transcription of other Bsa T3SS genes leaving the role of BprD as a regulator in doubt (48). More recently, a bprD mutant of B. pseudomallei K96243 was shown to have increased expression of bprC (80), a downstream gene in the same operon shown to be involved in regulation of the virulence-associated T6SS (81). The bprD mutant also showed decreased time to death in intraperitoneally inoculated BALB/c mice demonstrating its importance in vivo and indicating its possible role as a negative regulator of virulence (80). It is not without precedent that a regulator of the T3SS is also a substrate for secretion (e.g. LcrQ of Yersinia) (82).
Another candidate protein identified in this study, BPSS0860, appears to be a fliD homolog. There is a second copy of fliD (BPSL3320) located within the flagellar locus on Chromosome 1, which had protein abundance ratios that were similar between all strains. Interestingly, the expression of BPSL3320 was up-regulated, whereas the expression of BPSS0860 was down-regulated in the organs of hamsters infected by B. pseudomallei when compared with bacteria grown in vitro (83). Using our c-Myc expression system we were unable to establish Bsa-dependent secretion of BPSS0860; however, it is possible it may be coregulated with the Bsa T3SS in a similar manner to BPSS1512. One recent study has shown an effector protein may be secreted by more than one secretion system, thereby masking its secretion by the T3SS (84). This could also be the case for BPSS0860.
At the time of writing, little is known about the post-translational control of the Bsa T3SS. Generally it is accepted that T3SSs are under tight temporal regulation to ensure substrates are secreted in the correct order (17). This starts with assembly of the membrane-associated structural components, assembly of the needle with needle tip, secretion of translocators to form a pore in the host cell membrane, followed by secretion of effector proteins. Changes in the patterns of secreted translocators and effector proteins in different mutant strains can inform us of the possible role these proteins play in the hierarchy of secretion.
Inactivation of the needle tip protein of B. pseudomallei, BipD, shows a large increase of levels of both translocators and effectors in the culture supernatant, in agreement with data published for the homologous Shigella protein IpaD (46, 52). IpaD is the needle cap protein, acting to block secretion of effector proteins until host cell contact has taken place (85). Because of its high similarity in both sequence and structure (53) it is perhaps unsurprising that BipD would have a similar effect on the levels of substrates secreted by the Bsa T3SS. Interestingly there was a high ratio of BipB, but not BipC in the WT B. pseudomallei 10276 strain culture supernatant compared with 10276 bsaZ::pDM4 (Table I). In Shigella, IpaB is present in the needle tip in complex with IpaD, whereas IpaC is secreted later, only upon host cell contact or activation (86). With this in mind it is possible that growth in LB lacks the stimuli required for BipC secretion in WT B. pseudomallei 10276.
Here we present evidence that BsaP functions as a “gatekeeper” protein for effectors in a manner similar to the homologous T3SS proteins InvE/SepL/MxiC/YopN-TyeA (54–60). Members of this family of proteins function differently from each other, but are all thought to be important in allowing effectors to be secreted by the mature T3SS. Here we show that deletion of bsaP creates a phenotype in which effector proteins are hypersecreted and levels of translocators decrease. To fully understand BsaP in the context of the other members of the “gatekeeper” family of proteins, more work would be required to determine the molecular interactions of BsaP with other components of the Bsa T3SS.
This study is another example of the power of a combination of classic targeted mutagenesis and a gel-free quantitative proteomics approach, such as iTRAQ, to study a complex secretion system, providing insights into the involvement of certain proteins on the regulation of the T3SS itself. Two new substrates of the Bsa T3SS were revealed by the study, confirming the effector repertoire is more complex than reported so far.
Supplementary Material
Acknowledgments
We thank, from Dundee Cell Products, Paul Ajuh for technical advice, Cristina Vazquez-Martin and Amy Wheat for help with MS. We would also like to thank Andy Gill and Dominic Kurian at The Roslin Institute for their kind advice. This work was supported by Institute Strategic Grant funding from the BBSRC. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (87) via the PRIDE partner repository with the data set identifier PXD001656 and 10.6019/PXD001656.
Footnotes
Author contributions: C.W.V., K.J.C., M.P.S., and J.M.S. designed research; C.W.V. performed research; C.W.V., K.J.C., and J.M.S. analyzed data; C.W.V. and J.M.S. wrote the paper.
 This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S5 and Tables S1 to S4.
1 The abbreviations used are:
- T3SS
- Type III secretion system
- B. pseudomallei
- Burkholderia pseudomallei
- Bsa
- Burkholderia secretion apparatus
- iTRAQ
- Isobaric tags for relative and absolute quantification
- WT
- wild-type
- CHBP
- Cif homolog of Burkholderia pseudomallei
- IQR
- interquartile range
- Cm
- chloramphenicol
- Amp
- ampicillin
- Kan
- kanamycin
- Tet
- tetracycline
- EPEC
- enteropathogenic Escherichia coli
- MNGC
- multi-nucleated giant cell.
REFERENCES
- 1. Strauss J., Groves M., Mariappan M., Ellison D. W. (1969) Melioidosis in Malaysia II. Distribution of Pseudomonas pseudomallei in soil and surface water. Am. J. Trop. Med. Hyg. 18, 698–702 [PubMed] [Google Scholar]
- 2. Currie B. J., Ward L., Cheng A. C. (2010) The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl. Trop. Dis. 4, e900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiersinga W. J., Currie B. J., Peacock S. J. (2012) Melioidosis. N. Engl. J. Med. 367, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 4. Limmathurotsakul D., Wongratanacheewin S., Teerawattanasook N., Wongsuvan G., Chaisuksant S., Chetchotisakd P., Chaowagul W., Day N. P. J., Peacock S. J. (2010) Increasing incidence of human melioidosis in Northeast Thailand. Am. J. Trop. Med. Hyg. 82, 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiersinga W. J., van der Poll T., White N. J., Day N. P., Peacock S. J. (2006) Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282 [DOI] [PubMed] [Google Scholar]
- 6. Jones A. L., Beveridge T. J., Woods D. E. (1996) Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stevens M. P., Stevens J. M., Jeng R. L., Taylor L. A., Wood M. W., Hawes P., Monaghan P., Welch M. D., Galyov E. E. (2005) Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol. Microbiol. 56, 40–53 [DOI] [PubMed] [Google Scholar]
- 8. Kespichayawattana W., Rattanachetkul S., Wanun T., Utaisincharoen P., Sirisinha S. (2000) Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect. Immun. 68, 5377–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarz S., West T. E., Boyer F., Chiang W.-C., Carl M. A., Hood R. D., Rohmer L., Tolker-Nielsen T., Skerrett S. J., Mougous J. D. (2010) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6, e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. French C. T., Toesca I. J., Wu T.-H., Teslaa T., Beaty S. M., Wong W., Liu M., Schröder I., Chiou P.-Y., Teitell M. A., Miller J. F. (2011) Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. Proc. Natl. Acad. Sci. U.S.A. 108, 12095–12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwarz S., Singh P., Robertson J. D., LeRoux M., Skerrett S. J., Goodlett D. R., West T. E., Mougous J. D. (2014) VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toesca I. J., French C. T., Miller J. F. (2014) The Type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by Pseudomallei group Burkholderia species. Infect. Immun. 82, 1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galán J. E., Curtiss R. (1989) Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 86, 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maurelli A. T., Baudry B., D'Hautevil H., Hale T. L., Sansonettil P. J. (1985) Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gemski P., Lazere J. R., Casey T., Wohlhieter J. A. (1980) Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect. Immun. 28, 1044–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jarvis K. G., Girón J. a, Jerse a E., McDaniel T. K., Donnenberg M. S., Kaper J. B. (1995) Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U.S.A. 92, 7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Büttner D. (2012) Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dean P. (2011) Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35, 1100–1125 [DOI] [PubMed] [Google Scholar]
- 19. Rainbow L., Hart C. A., Winstanley C. (2002) Distribution of type III secretion gene clusters in Burkholderia pseudomallei, B. thailandensis, and B. mallei. J. Med. Microbiol. 51, 374–384 [DOI] [PubMed] [Google Scholar]
- 20. Winstanley C., Hales B., Hart C. (1999) Evidence for the presence in Burkholderia pseudomallei of a type III secretion system-associated gene cluster. J. Med. Microbiol. 48, 649–656 [DOI] [PubMed] [Google Scholar]
- 21. Stevens M. P., Wood M. W., Taylor L. A., Monaghan P., Hawes P., Jones P. W., Wallis T. S., Galyov E. E. (2002) An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behavior of the pathogen. Mol. Microbiol. 46, 649–659 [DOI] [PubMed] [Google Scholar]
- 22. Stevens M. P., Haque A., Atkins T., Hill J., Wood M. W., Easton A., Nelson M., Underwood-Fowler C., Titball R. W., Bancroft G. J., Galyov E. E. (2004) Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150, 2669–2676 [DOI] [PubMed] [Google Scholar]
- 23. Warawa J., Woods D. E. (2005) Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242, 101–108 [DOI] [PubMed] [Google Scholar]
- 24. Burtnick M. N., Brett P. J., Nair V., Warawa J. M., Woods D. E., Gherardini F. C. (2008) Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect. Immun. 76, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muangman S., Korbsrisate S., Muangsombut V., Srinon V., Adler N. L., Schroeder G. N., Frankel G., Galyov E. E. (2011) BopC is a type III secreted effector protein of Burkholderia pseudomallei. FEMS Microbiol. Lett. 323, 75–82 [DOI] [PubMed] [Google Scholar]
- 26. Stevens M. P., Friebel A., Taylor L. A., Wood M. W., Brown P. J., Hardt W.-D., Galyov E. E. (2003) A Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity. J. Bacteriol. 185, 4992–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pumirat P., Broek C. Vander, Juntawieng N., Muangsombut V., Kiratisin P., Pattanapanyasat K., Stevens J. M., Stevens M. P., Korbsrisate S. (2014) Analysis of the prevalence, secretion, and function of a cell cycle-inhibiting factor in the melioidosis pathogen Burkholderia pseudomallei. PLoS One 9, e96298. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Srinon V., Muangman S., Imyaem N., Muangsombut V., Lazar Adler N. R., Galyov E. E., Korbsrisate S. (2013) Comparative assessment of the intracellular survival of the Burkholderia pseudomallei bopC mutant. J. Microbiol. 51, 522–526 [DOI] [PubMed] [Google Scholar]
- 29. Marchès O., Ledger T. N., Boury M., Ohara M., Tu X., Goffaux F., Mainil J., Rosenshine I., Sugai M., De Rycke J., Oswald E. (2003) Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 30. Gong L., Cullinane M., Treerat P., Ramm G., Prescott M., Adler B., Boyce J. D., Devenish R. J. (2011) The Burkholderia pseudomallei type III secretion system and BopA are required for evasion of LC3-associated phagocytosis. PLoS One 6, e17852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Whitlock G. C., Estes D. M., Young G. M., Young B., Torres A. G. (2008) Construction of a reporter system to study Burkholderia mallei type III secretion and identification of the BopA effector protein function in intracellular survival. Trans. R. Soc. Trop. Med. Hyg. 102, S127–S133 [DOI] [PubMed] [Google Scholar]
- 32. Dean P., Kenny B. (2009) The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12, 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suwannasaen D., Mahawantung J., Chaowagul W., Limmathurotsakul D., Felgner P. L., Davies H., Bancroft G. J., Titball R. W., Lertmemongkolchai G. (2011) Human immune responses to Burkholderia pseudomallei characterized by protein microarray analysis. J. Infect. Dis. 203, 1002–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tippayawat P., Pinsiri M., Rinchai D., Riyapa D., Romphruk A., Gan Y.-H., Houghton R. L., Felgner P. L., Titball R. W., Stevens M. P., Galyov E. E., Bancroft G. J., Lertmemongkolchai G. (2011) Burkholderia pseudomallei proteins presented by monocyte-derived dendritic cells stimulate human memory T cells in vitro. Infect. Immun. 79, 305–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Felgner P. L., Kayala M. A., Vigil A., Burk C., Nakajima-Sasaki R., Pablo J., Molina D. M., Hirst S., Chew J. S. W., Wang D., Tan G., Duffield M., Yang R., Neel J., Chantratita N., Bancroft G., Lertmemongkolchai G., Davies D. H., Baldi P., Peacock S., Titball R. W. (2009) A Burkholderia pseudomallei protein microarray reveals serodiagnostic and cross-reactive antigens. Proc. Natl. Acad. Sci. U.S.A. 106, 13499–13504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tippayawat P., Saenwongsa W., Mahawantung J., Suwannasaen D., Chetchotisakd P., Limmathurotsakul D., Peacock S. J., Felgner P. L., Atkins H. S., Titball R. W., Bancroft G. J., Lertmemongkolchai G. (2009) Phenotypic and functional characterization of human memory T cell responses to Burkholderia pseudomallei. PLoS Negl. Trop. Dis. 3, e407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heine S. J., Diaz-McNair J., Martinez-Becerra F. J., Choudhari S. P., Clements J. D., Picking W. L., Pasetti M. F. (2013) Evaluation of immunogenicity and protective efficacy of orally delivered Shigella type III secretion system proteins IpaB and IpaD. Vaccine 31, 2919–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez-Becerra F. J., Kissmann J. M., Diaz-McNair J., Choudhari S. P., Quick A. M., Mellado-Sanchez G., Clements J. D., Pasetti M. F., Picking W. L. (2012) Broadly protective Shigella vaccine based on type III secretion apparatus proteins. Infect. Immun. 80, 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Williamson E. D., Eley S. M., Stagg A. J., Green M., Russell P., Titball R. W. (1997) A subunit vaccine elicits IgG in serum, spleen cell cultures and bronchial washings and protects immunized animals against pneumonic plague. Vaccine 15, 1079–1084 [DOI] [PubMed] [Google Scholar]
- 40. Cornelius C. A., Quenee L. E., Overheim K. A., Koster F., Brasel T. L., Elli D., Ciletti N. A., Schneewind O. (2008) Immunization with recombinant V10 protects cynomolgus macaques from lethal pneumonic plague. Infect. Immun. 76, 5588–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kurtz J. R., Petersen H. E., Frederick D. R., Morici L. A., McLachlan J. B. (2014) Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect. Immun. 82, 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee V. T., Mazmanian S. K., Schneewind O. (2001) A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J. Bacteriol. 183, 4970–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olive A. J., Kenjale R., Espina M., Moore D. S., Picking W. L., Picking W. D. (2007) Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect. Immun. 75, 2626–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michiels T., Wattiau P., Brasseur R., Ruysschaert J. M., Cornelis G. (1990) Secretion of Yop proteins by Yersiniae. Infect. Immun. 58, 2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Forsberg Å., Wolf-Watz H. (1988) The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-plB1-encoded trans-acting elements controlled by temperature and calcium. Mol. Microbiol. 2, 121–133 [DOI] [PubMed] [Google Scholar]
- 46. Parsot C., Ménard R., Gounon P., Sansonetti P. J. (1995) Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16, 291–300 [DOI] [PubMed] [Google Scholar]
- 47. Jamison W. P., Hackstadt T. (2008) Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb. Pathog. 45, 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun G. W., Chen Y., Liu Y., Tan G.-Y. G., Ong C., Tan P., Gan Y.-H. (2010) Identification of a regulatory cascade controlling Type III Secretion System 3 gene expression in Burkholderia pseudomallei. Mol. Microbiol. 76, 677–689 [DOI] [PubMed] [Google Scholar]
- 49. Pumirat P., Saetun P., Sinchaikul S., Chen S.-T., Korbsrisate S., Thongboonkerd V. (2009) Altered secretome of Burkholderia pseudomallei induced by salt stress. Biochim. Biophys. Acta 1794, 898–904 [DOI] [PubMed] [Google Scholar]
- 50. Deng W., Yu H. B., de Hoog C. L., Stoynov N., Li Y., Foster L. J., Finlay B. B. (2012) Quantitative proteomic analysis of type III secretome of enteropathogenic Escherichia coli reveals an expanded effector repertoire for attaching/effacing bacterial pathogens. Mol. Cell. Proteomics 11, 692–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deng W., de Hoog C. L., Yu H. B., Li Y., Croxen M. A., Thomas N. A., Puente J. L., Foster L. J., Finlay B. B. (2010) A comprehensive proteomic analysis of the type III secretome of Citrobacter rodentium. J. Biol. Chem. 285, 6790–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Picking W. L., Nishioka H., Hearn P. D., Baxter M. A., Harrington A. T., Blocker A., Picking W. D. (2005) IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect. Immun. 73, 1432–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Erskine P. T., Knight M. J., Ruaux A., Mikolajek H., Wong Fat Sang N., Withers J., Gill R., Wood S. P., Wood M., Fox G. C., Cooper J. B. (2006) High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei. J. Mol. Biol. 363, 125–136 [DOI] [PubMed] [Google Scholar]
- 54. Kubori T., Galán J. E. J. (2002) Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J. Bacteriol. 184, 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kresse A., Beltrametti F., Müller A. (2000) Characterization of SepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 182, 6490–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deng W., Puente J. L., Gruenheid S., Li Y., Vallance B. A., Vázquez A., Barba J., Ibarra J. A., O'Donnell P., Metalnikov P., Ashman K., Lee S., Goode D., Pawson T., Finlay B. B. (2004) Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deng W., Li Y., Hardwidge P. R., Elizabeth A., Pfuetzner R. A., Lee S., Strynakda N. C. J., Puente J. L., Frey E. A., Gruenheid S., Finlay B. B. (2005) Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Botteaux A., Sory M. P., Biskri L., Parsot C., Allaoui A. (2009) MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 71, 449–460 [DOI] [PubMed] [Google Scholar]
- 59. Forsberg Å., Viitanen A. M., Skurnik M., Wolf-Watz H. (1991) The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5, 977–986 [DOI] [PubMed] [Google Scholar]
- 60. Iriarte M., Sory M. P., Boland A., Boyd A. P., Mills S. D., Lambermont I., Cornelis G. R. (1998) TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 17, 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Allaoui A., Woestyn S., Sluiters C., Cornelis G. R. (1994) YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J. Bacteriol. 176, 4534–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Logue C.-A., Peak I. R. A., Beacham I. R. (2009) Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates. J. Microbiol. Methods 76, 320–323 [DOI] [PubMed] [Google Scholar]
- 63. Ali S., Steinkasserer A. (1995) PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18, 746–750 [PubMed] [Google Scholar]
- 64. Choi K.-H., Kumar A., Schweizer H. P. (2006) A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64, 391–397 [DOI] [PubMed] [Google Scholar]
- 65. Caldwell R. B., Lattemann C. T. (2004) Simple and reliable method to precipitate proteins from bacterial culture supernatant. Appl. Environ. Microbiol. 70, 610–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Boisvert F.-M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Olsen J. V., de Godoy L. M. F., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 68. Holden M. T. G., Titball R. W., Peacock S. J., Cerdeño-Tárraga A. M., Atkins T., Crossman L. C., Pitt T., Churcher C., Mungall K., Bentley S. D., Sebaihia M., Thomson N. R., Bason N., Beacham I. R., Brooks K., Brown K. A., Brown N. F., Challis G. L., Cherevach I., Chillingworth T., Cronin A., Crossett B., Davis P., DeShazer D., Feltwell T., Fraser A., Hance Z., Hauser H., Holroyd S., Jagels K., Keith K. E., Maddison M., Moule S., Price C., Quail M. A., Rabbinowitsch E., Rutherford K., Sanders M., Simmonds M., Songsivilai S., Stevens K., Tumapa S., Vesaratchavest M., Whitehead S., Yeats C., Barrell B. G., Oyston P. C. F., Parkhill J. (2004) Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. U.S.A. 101, 14240–14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harding S. V., Sarkar-Tyson M., Smither S. J., Atkins T. P., Oyston P. C. F., Brown K. A., Liu Y., Wait R., Titball R. W. (2007) The identification of surface proteins of Burkholderia pseudomallei. Vaccine 25, 2664–2672 [DOI] [PubMed] [Google Scholar]
- 70. Schell M. a, Zhao P., Wells L. (2011) Outer membrane proteome of Burkholderia pseudomallei and Burkholderia mallei from diverse growth conditions. J. Proteome Res. 10, 2417–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Su Y.-C., Wan K.-L., Mohamed R., Nathan S. (2008) A genome level survey of Burkholderia pseudomallei immunome expressed during human infection. Microbes Infect. 10, 1335–1345 [DOI] [PubMed] [Google Scholar]
- 72. Burtnick M. N., Brett P. J., DeShazer D. (2014) Proteomic analysis of the Burkholderia pseudomallei type II secretome reveals hydrolytic enzymes, novel proteins, and the deubiquitinase TssM. Infect. Immun. 82, 3214–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ooi W. F., Ong C., Nandi T., Kreisberg J. F., Chua H. H., Sun G. W., Chen Y., Mueller C., Conejero L., Eshaghi M., Ang R. M. L., Liu J., Sobral B. W., Korbsrisate S., Gan Y.-H., Titball R. W., Bancroft G. J., Valade E., Tan P. (2013) The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 9, e1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schumacher J., Waite C. J., Bennett M. H., Perez M. F., Shethi K., Buck M. (2014) Differential secretome analysis of Pseudomonas syringae pv tomato using gel-free MS proteomics. Front. Plant Sci. 5, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kurushima J., Kuwae A., Abe A. (2012) The type III secreted protein BspR regulates the virulence genes in Bordetella bronchiseptica. PLoS One 7, e38925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vanden Bergh P., Heller M., Braga-Lagache S., Frey J. (2013) The Aeromonas salmonicida subsp. salmonicida exoproteome: determination of the complete repertoire of Type-Three Secretion System effectors and identification of other virulence factors. Proteome Sci. 11, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shanks J., Burtnick M. N., Brett P. J., Waag D. M., Spurgers K. B., Ribot W. J., Schell M. a, Panchal R. G., Gherardini F. C., Wilkinson K. D., Deshazer D. (2009) Burkholderia mallei tssM encodes a putative deubiquitinase that is secreted and expressed inside infected RAW 264.7 murine macrophages. Infect. Immun. 77, 1636–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tan K. S., Chen Y., Lim Y.-C., Tan G.-Y. G., Liu Y., Lim Y.-T., Macary P., Gan Y.-H. (2010) Suppression of host innate immune response by Burkholderia pseudomallei through the virulence factor TssM. J. Immunol. 184, 5160–5171 [DOI] [PubMed] [Google Scholar]
- 79. Burtnick M. N., Brett P. J., Harding S. V., Ngugi S. A., Ribot W. J., Chantratita N., Scorpio A., Milne T. S., Dean R. E., Fritz D. L., Peacock S. J., Prior J. L., Atkins T. P., Deshazer D. (2011) The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chirakul S., Bartpho T., Wongsurawat T., Taweechaisupapong S., Karoonutaisiri N., Talaat A. M., Wongratanacheewin S., Ernst R. K., Sermswan R. W. (2014) Characterization of BPSS1521 (bprD), a regulator of Burkholderia pseudomallei virulence gene expression in the mouse model. PLoS One 9, e104313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen Y., Wong J., Sun G. W., Liu Y., Tan G.-Y. G., Gan Y.-H. (2011) Regulation of Type VI Secretion System during Burkholderia pseudomallei infection. Infect. Immun. 79, 3064–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cambronne E. D., Cheng L. W., Schneewind O. (2000) LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 37, 263–273 [DOI] [PubMed] [Google Scholar]
- 83. Tuanyok A., Tom M., Dunbar J., Woods D. E. (2006) Genome-wide expression analysis of Burkholderia pseudomallei infection in a hamster model of acute melioidosis. Infect. Immun. 74, 5465–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Neeld D., Jin Y., Bichsel C., Jia J., Guo J., Bai F., Wu W., Ha U.-H., Terada N., Jin S. (2014) Pseudomonas aeruginosa injects NDK into host cells through a type III secretion system. Microbiology 160, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Roehrich A. D., Guillossou E., Blocker A. J., Martinez-Argudo I. (2013) Shigella IpaD has a dual role: signal transduction from the type III secretion system needle tip and intracellular secretion regulation. Mol. Microbiol. 87, 690–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Epler C. R., Dickenson N. E., Olive A. J., Picking W. L., Picking W. D. (2009) Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect. Immun. 77, 2754–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N., Binz P.A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R.J., Kraus H.J., Albar J.P., Martinez-Bartolomé S., Apweiler R., Omenn G.S., Martens L., Jones A.R., Hermjakob H. (2014) ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nature Biotechnol. 30, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.