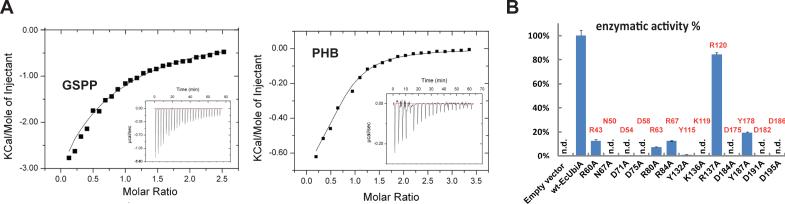
Figure 3. Binding and activity assays of UbiA enzymes.
(A) ITC measurements of the GSPP (left) and PHB (right) binding to ApUbiA in presence of Mg. (B) Prenyltransferase activities of EcUbiA mutants. Conserved residues at the central cavity are mutated (n.d.: not detected; error bars: s.e.m. of duplicated assays). Corresponding residues in the ApUbiA structure (Fig. 2A) are labeled in red. R137 is a positive control that is not at the active site in our structure, yet was predicted important in an in silico model (19).