Abstract
The normal expression of β-globin protein in mature erythrocytes is critically dependent on post-transcriptional events in erythroid progenitors that ensure the high stability of β-globin mRNA. Previous work has revealed that these regulatory processes require AUF-1 and YB-1, two RNA-binding proteins that assemble an mRNP β-complex on the β-globin 3′UTR. Here, we demonstrate that the β-complex organizes during the erythropoietic interval when both β-globin mRNA and protein accumulate rapidly, implicating the importance of this regulatory mRNP to normal erythroid differentiation. Subsequent functional analyses link β-complex assembly to the half-life of β-globin mRNA in vivo, providing a mechanistic basis for this regulatory activity. AUF-1 and YB-1 appear to serve a redundant post-transcriptional function, as both β-complex assembly and β-globin mRNA levels are reduced by coordinate depletion of the two factors, and can be restored by independent rescue with either factor alone. Additional studies demonstrate that the β-complex assembles more efficiently on polyadenylated transcripts, implicating a model in which the β-complex enhances the binding of PABPC1 to the poly(A) tail, inhibiting mRNA deadenylation and consequently effecting the high half-life of β-globin transcripts in erythroid progenitors. These data specify a post-transcriptional mechanism through which AUF1 and YB1 contribute to the normal development of erythropoietic cells, as well as to non-hematopoietic tissues in which AUF1-and YB1-based regulatory mRNPs have been observed to assemble on heterologous mRNAs.
Keywords: Erythroid development, β globin, mRNA stability
1. Introduction
Post-transcriptional events that regulate the stabilities of individual mRNAs are increasingly recognized for their impact on cell development and differentiation: genome-wide analyses attribute ~50% of changes in gene expression to processes that alter mRNA half-lives (Cheadle et al., 2005). Events that regulate mRNA stability are particularly important in terminally differentiating erythroid progenitor cells that are transcriptionally silent, but remain translationally active (Greer et al., 2009). The critical impact of post-transcriptional process to normal erythropoiesis is illustrated by functional studies of mRNAs that encode human α and β globins, the principal proteins expressed in erythrocytes. The two globin mRNAs exhibit half-life values that exceed 20 hr in vivo (Ross and Sullivan, 1985; Volloch and Housman, 1981), which provide for their selective enrichment both in transcriptionally silent erythroid progenitors and in their anucleate progeny, and account for the consequent high-level expression of their encoded α-and β-globin proteins. Congenital molecular defects that compromise the stabilities of either mRNA effect significant decrements in globin production that can precipitate serious clinical conditions that are collectively termed thalassemias (Clegg et al., 1971; Peixeiro et al., 2011; Weiss and Liebhaber, 1994). The clear relevance of post-transcriptional processes to normal hemoglobinization has fostered intense interest in defining the cis-acting mRNA determinants and corresponding trans-acting factors that mediate this critical aspect of definitive erythropoiesis.
We recently identified two trans-acting factors that regulate the level of β-globin mRNA in erythroid cells through a mechanism that, while not yet fully elucidated, exhibits several novel features (van Zalen et al., 2012). The two RNA-binding proteins (RBPs), AUF-1 and YB-1, assemble an erythroidspecific messenger ribonucleoprotein (mRNP) on a region of 3′UTR that has previously been implicated as a determinant of β-globin mRNA stability (Jiang et al., 2006). This mRNP, which we term the ‘β-complex’, is cytoplasm-restricted and erythroidspecific, suggesting that it participates in regulating β-globin mRNA at the stages of erythroid differentiation when progenitor cells are transcriptionally silenced and, subsequently, extrude their nuclei. Several well-described properties of AUF-1 and YB-1 are consistent with this possibility: both proteins express at high levels in late erythropoiesis (van Zalen et al., 2012), display mRNA-stabilizing activities (Capowski et al., 2001; Fellows et al., 2012; McGray et al., 2011), and interact with poly(A) binding protein (PABP) (Higashi et al., 2011; Laroia et al., 1999), a factor that enhances the half-lives of heterologous mRNAs by maintaining the integrities of their polyadenylate tails (Bernstein et al., 1989; Wilusz et al., 2001).
Investigations from our laboratory and from others have provided insights into the specific mechanisms through which AUF-1 and YB-1 may effect the unusually high stability of β-globin mRNA. Functional analyses conducted in vivo suggest that AUF-1 and YB-1 act redundantly, as coordinate depletion of both factors is required to reduce levels of β-globin mRNA at steady state (van Zalen et al., 2012). Structural assays performed in vitro demonstrate that the two proteins bind independently to the β-globin 3′UTR, fully according with the proposed model for functional redundancy (van Zalen et al., 2012). While revealing, these data do not address more fundamental mechanistic questions, e.g., whether AUF-1 and YB-1 bind simultaneously to single β-globin transcripts or, alternatively, bind independently to different β-globin mRNAs. Both possibilities are consistent with previously described interactions between the two RBPs and several non-globin RNAs. For example, AUF-1 and YB-1 bind simultaneously to a synthetic AU-rich RNA (Moraes et al., 2003), consistent with their inclusion in a functional multiprotein mRNP (Skalweit et al., 2003). In contrast, AUF-1 and YB-1 bind independently to defined mRNA-stability motifs in the 3′UTRs of mRNAs that encode GMCSF, VEGF, and TSP-1 (Capowski et al., 2001; Fellows et al., 2012; McGray et al., 2011), illustrating the capacities of these regulatory factors for structural and functional independence. A more comprehensive description of the mechanism(s) through which AUF-1 and YB-1 regulate β-globin mRNA, as either heteromeric or monomeric mRNPs, is fundamental to understanding the post-transcriptional processes that are required for normal erythropoiesis.
It is likely that the mechanism through which AUF-1 and YB-1 regulate levels of β-globin mRNA reflects a modification of molecular processes that contribute to the decay of heterologous transcripts. mRNA degradation in eukaryotes characteristically initiates with 3′→5′ exonucleolytic digestion of the poly(A) tail, proceeds with enzymatic removal of the 5′m7GpppG cap structure, and is completed by endonucleolytic degradation of the deadenylated, decapped transcript (Parker and Song, 2004; Wahle and Winkler, 2013). This process is inhibited by the cytoplasmic form of PABP (PABPC1), which binds as a multimer to the mRNA poly(A) tail and protects it from decay-initiating deadenylation (Bernstein et al., 1989; Wilusz et al., 2001). A number of RBPs that stabilize heterologous mRNAs – including αCP, HuR, and TTP – appear to execute their effects by enhancing the interaction between PABPC1 and the poly(A) tail (Kedar et al., 2010; Nagaoka et al., 2006; Wang et al., 1999). As both AUF-1 and YB-1 have previously been shown to interact with PABPC1 bound to non-globin mRNAs (Higashi et al., 2011; Laroia et al., 1999), it is reasonable to speculate that these two regulatory factors stabilize β-globin mRNA through a related process.
The present manuscript defines key features of the mechanism through which AUF-1 and YB-1 regulate β-globin mRNA during normal erythropoiesis. We demonstrate that primary erythroid progenitor cells acquire the capacity to assemble an mRNP β-complex during the developmental interval when there is both an exponential increase in the level of β-globin mRNA and a corresponding accumulation of β-globin protein. Subsequent experiments demonstrate that the two components of the β-complex – AUF-1 and YB-1 – exhibit both structural and functional redundancy. Additional analyses consider the mechanism that underlies the mRNA-stabilizing properties of the β-complex, and establish causal relationships between β-complex assembly, β-globin mRNA polyadenylation, and β-globin mRNA stability. Our results are consistent with the unique process of global transcriptional silencing that characterizes terminal differentiation in erythroid cells, and demonstrate that the β-complex ensures the high half-life of β-globin mRNA by enhancing a protective interaction between PABPC1 and the mRNA poly(A) tail. The data additionally specify a post-transcriptional mechanism that may participate in the regulation of mRNAs that assemble AUF1-and YB1-based mRNPs in a variety of non-hematopoietic cells and tissues.
2. Results
2.1. Concurrent induction of β-globin expression and β-complex assembly during erythropoiesis
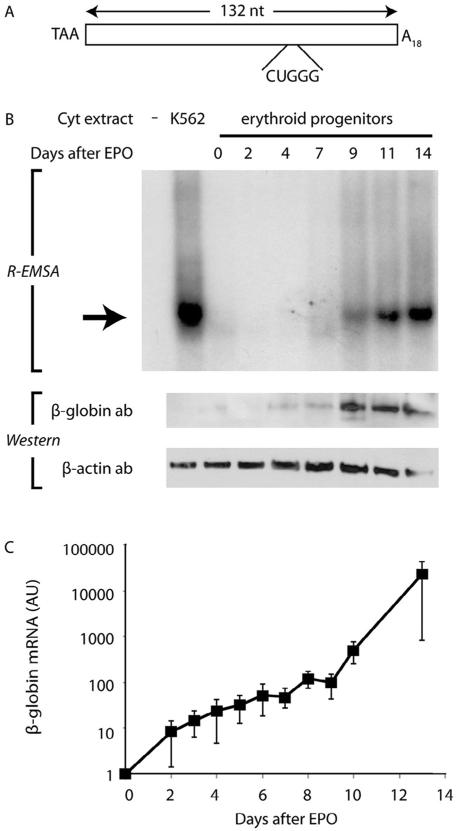
The putative regulatory properties of the mRNP β-complex predict its assembly in erythroid progenitor cells during the interval when β-globin mRNA begins to accumulate rapidly. We previously validated the lineage specificity of this expectation by demonstrating that the β-complex forms during erythroid, but not granulocytic-monocytic differentiation (van Zalen et al., 2012). To define the temporal characteristics of this process, we assessed the capacity of hematopoietic progenitor cells to assemble a β-complex at defined intervals following their induction to erythroid differentiation. RNA electromobility shift assays (R-EMSAs), using a RNA probe comprising the full-length β-globin 3′UTR (Fig. 1A), revealed that erythroid-differentiated CD34+ cells initiate β-complex formation approximately 9 d after addition of EPO, coincident with their induction of β-globin protein to high-level expression (Fig. 1B). As might be anticipated, the rapid accumulation of β globin in these cells is accompanied by an exponential increase in β-globin mRNA, which parallels the increase in β-complex assembly and is consistent with the proposed mRNA-regulatory properties of the β-complex (Fig. 1C). The capacity of erythroid progenitor cells to assemble a β-complex during this interval – a characteristic that is progressively enhanced as differentiation proceeds – implicates its importance to the selective enrichment of β-globin mRNA.
Fig. 1.
Assembly of the β-complex and coordinate induction of β-globin expression during erythropoiesis. (A) Schematic depiction of the 132-nt in vitro transcribed [32P]-labeled RNA, corresponding to the full-length β-globin 3′UTR, that is used for all EMSAs. The translation termination codon (TAA), 18-nt poly(A) tail (A18), and 5-nt β-complex target site (CUGGG) are indicated. (B) β-Globin protein accumulates in primary erythroid cells that support β-complex formation. Top: R-EMSAs were conducted on a [32P]-labeled β-globin RNA 3′UTR using cytoplasmic extracts purified from human EPO-induced CD34+ cells at defined intervals. K562 cytoplasmic extract was used as a positive control. The β-complex is indicated by an arrow. Bottom: Parallel Western-transfer analyses of cytoplasmic extracts for β-globin and control β-actin expression. (C) β-Complex assembly in erythroid differentiation parallels an exponential increase in β-globin mRNA. RT-qPCR analyses were conducted on RNA isolated from human EPO-induced CD34+ erythroid progenitor cells. Each point represents the mean±SEM values from at least 4 biological replicates, normalized to values for endogenous control β-actin and GAPDH mRNAs. The normalized level of β-globin mRNA at day 0 was arbitrarily assigned unit value. Note logarithmic ordinate scale.
2.2. The β-complex regulates the cytoplasmic half-life of β-globin mRNA
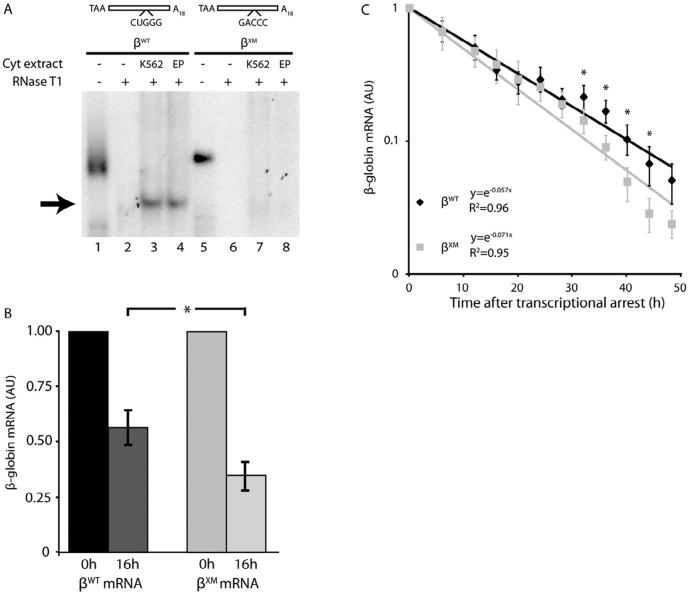
Previous analyses conducted in erythroid K562 cells indicate that the β-complex targets a guanosine-rich pentanucleotide at positions 65–69 of the β-globin 3′UTR (Sup Fig. S1) (van Zalen et al., 2012). We confirmed this structural determinant of β-complex assembly – in a biologically meaningful context – by conducting R-EMSAs using cytoplasmic extracts from EPO-induced human CD34+ cells. The site-specific CUGGG→GACCC mutation fully ablated β-complex assembly in extracts prepared from both erythroid-phenotype K562 cells and primary erythroid progenitor cells (Fig. 2A). This result validates the region of 3′UTR that participates in β-complex assembly, demonstrates that it functions as a target for regulatory trans-acting factors in primary erythroid cells, and indicates that β-globin mRNAs carrying an ablative pentanucleotide mutation (βXM) can be used to investigate the function of the β-complex in cultured K562 cells (Rutherford et al., 1981).
Fig. 2.
The half-life of β-globin mRNA is reduced by a 3′UTR mutation that ablates assembly of the mRNP β-complex. (A) The β-complex assembles on a site-specific region of β-globin 3′UTR in primary erythroid progenitor cells. R-EMSAs were conducted on β-globin 3′UTRs that contain either the wild-type sequence (βWT) or a site-specific CUGGG→GACCC mutation (βXM), using cytoplasmic extracts purified from both cultured K562 and primary human erythroid progenitor (EP) cells. Undigested probes (20%) were assessed in parallel to control for transcript integrities and intensities (lanes 1, 5). An arrow indicates the β-complex. (B) A mutation in the mRNA-binding site for the β-complex reduces the accumulation of β-globin mRNA in cultured erythroid cells. K562/tTA cells were transiently transfected with constructs that conditionally transcribe either βWT or βXM mRNAs, along with a construct that expresses a control luciferase mRNA. β-Globin gene transcription was induced for 4 hr, then silenced by addition of dox to the culture medium. Levels of βWT or βXM mRNAs were quantified 0 and 16 hr after transcriptional arrest, then normalized to levels of control dox-indifferent luciferase mRNA. The normalized value for each mRNA at time=0 hr was arbitrarily assigned unit value. Each bar depicts the mean±SEM values for 5 independent biological replicates. * p < 0.05 (one-tailed paired t-test for βWT v βXM values). (C) The XM mutation destabilizes β-globin mRNA in erythroid cells. K562/tTA cells were stably transfected with constructs encoding βWT or βXM mRNAs, and pooled cells subsequently selected with hygromycin. The half-life value for each mRNA was assessed by quantifying its levels at defined intervals following gene-specific transcriptional arrest, and normalizing these values to the levels of dox-indifferent β-actin mRNA. The normalized value for each mRNA at time=0 hr was arbitrarily assigned unit value. Each point depicts the mean±SEM values for 6 independent biological replicates. Half-life values were calculated by regressing the data to a first-order exponential decay function. The coefficient of determination (R2) for each decay function is indicated. *p < 0.05 (one-tailed paired t-test for βWT v βXM values).
The cytoplasm-restricted assembly of the β-complex (van Zalen et al., 2012) and the known RNA-regulatory properties of its constituent trans-acting factors (Capowski et al., 2001; Fellows et al., 2012; McGray et al., 2011) predict that the mRNP participates in post-transcriptional processes affecting the stability and/or the translational efficiency of mature β-globin mRNA. Our previous studies demonstrated a significant defect in the accumulation of β-globin mRNA when the integrities of either the cis- or the trans-acting elements of the β-complex were interrupted (Jiang et al., 2006; van Zalen et al., 2012), suggesting that the β-complex is integral to β-globin post-transcriptional regulatory mechanisms. To test this possibility, K562/tTA cells were transiently transfected with doxycycline (dox)-conditional genes encoding β-globin mRNAs that do (βWT) or do not (βXM) assemble a β-complex. Dox exposure specifically arrests β-globin gene transcription in these cells, permitting the contribution of post-mechanisms to the regulated accumulation of β-globin mRNA to be accurately assessed. Serial analyses revealed that βXM mRNA accumulates to a significantly lower level compared to βWT mRNA following a 16-hr interval of transcriptional silence (Fig. 2B), validating the importance of the β-complex to β-globin mRNA half-life in vivo. We formally confirmed this possibility using K562/tTA cells engineered to stably express either βWT or βXM mRNAs, facilitating formal half-life (t1/2) analyses of their encoded mRNAs using a related transcriptional chase strategy (Fig. 2C). Replicate analyses yielded a t1/2 value of 9.3 hr for βXM mRNA, significantly lower than the t1/2 value of 13.4 hr observed for β WT mRNA. The ~4-hr decline in the half-life of βXM mRNA and the corresponding ~40% decrease in its accumulation concur in demonstrating the importance of the β-complex to the cytoplasmic stability of β-globin mRNA, and accord with expectations that we initially based upon both the cellular localization of this regulatory mRNP (van Zalen et al., 2012) and the timing of its assembly (Fig. 1). Collectively, these analyses indicate that the primary function of the β-complex in erythropoiesis is to maintain the high cytoplasmic stability of β-globin mRNA.
2.3. AUF-1 and YB-1 independently regulate β-globin mRNA levels
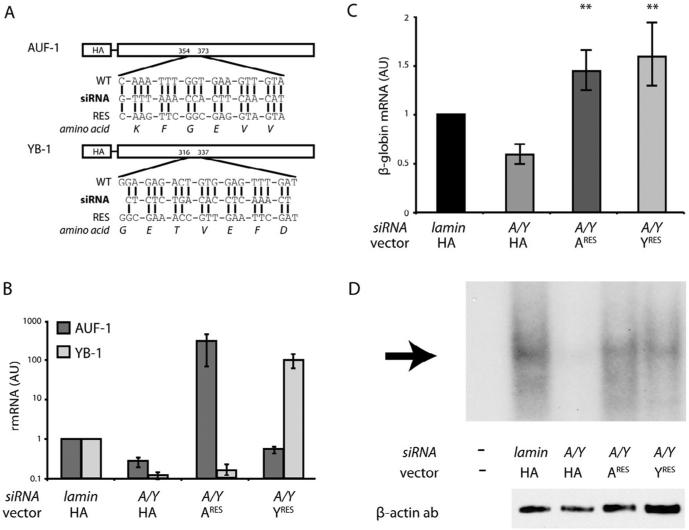
The level of cytoplasmic β-globin mRNA can be reduced in erythroid cells by coordinate depletion of AUF-1 and YB-1, but not by their independent knock-down, suggesting a functional redundancy in AUF-1 and YB-1 activity (van Zalen et al., 2012). To confirm this observation, we tested whether the reduced level of β-globin mRNA that results from coordinate depletion of AUF-1 and YB-1 could be restored to normal by exogenous expression of either regulatory factor alone. Our strategy employed HA-tagged AUF-1 and HA-tagged YB-1, each encoded by an mRNA containing synonymous codons that confer siRNA resistance, but do not alter the native amino-acid sequence (Fig. 3A, Sup Fig. S2). Endogenous levels of both AUF-1 and YB-1 mRNAs were significantly reduced in K562 cells transfected with the corresponding siRNAs, but were effectively substituted by siRNA-resistant exogenous HA-AUF-1 or HA-YB-1 mRNAs, respectively (Fig. 3B). The independent expression of either HA-AUF-1 or HA-YB-1 had a clear effect on the accumulation of β-globin mRNA in cells in which endogenous AUF-1 and YB-1 were coordinately depleted (Fig. 3C). Levels of β-globin mRNA were reduced by approximately one-half in AUF-1/YB-1-depleted K562 cells – relative to lamin siRNA-transfected control cells – but were restored above baseline in similarly depleted cells that independently expressed either siRNA-resistant HA-AUF-1 or siRNA-resistant HA-YB-1. For each experimental condition, R-EMSAs conducted on cytoplasmic extracts demonstrated a β-complex whose intensity paralleled the level of β-globin mRNA (Fig. 3D), consistent with earlier experiments showing that AUF-1 and YB-1 bind independently to the β-globin 3′UTR and assemble mRNPs with equivalent functions (van Zalen et al., 2012). These analyses confirm both structural and functional redundancy for AUF-1 and YB-1, since each can independently assemble a β-complex on the β-globin 3′UTR, and each can independently ensure high-level accumulation of β-globin mRNA.
Fig. 3.
Rescue expression of either AUF-1 or YB-1 independently restores β-globin mRNA to normal levels. (A) Sequence comparison of endogenous and siRNA-resistant HA-AUF-1 and HA-YB-1. For each factor, the siRNA sequence (bold), the endogenous mRNA target sequence (WT), the siRNA-resistant sequence (RES), and the translated amino-acid sequence are illustrated. Vertical lines indicate nucleotide identity. Numbers indicate the position of each sequence in the respective ORF. (B) Levels of siRNA-resistant HA-AUF-1 or HA-YB-1 mRNAs in siRNA-transfected and control cells. K562 cells were co-transfected with siRNAs targeting AUF-1 (A) and YB-1 (Y), along with rescue vectors encoding either siRNA-resistant HA-AUF-1 mRNA (ARES) or siRNA-resistant HA-YB-1 mRNA (YRES). AUF-1 and YB-1 mRNAs were quantified by RT-qPCR and normalized to average levels of control GAPDH and β-actin mRNAs. As a negative control, a vector encoding parental HA mRNA was co-transfected with an siRNA targeting lamin mRNA. Levels of AUF-1 and YB-1 in lamin siRNA-transfected cells were assigned unit value. Each bar represents the mean±SEM values for 5 experimental replicates. Note logarithmic ordinate scale. (C) Rescue of β-globin mRNA levels by siRNA-resistant HA-AUF-1 and siRNA-HA-YB-1. For experimental set-up, see panel B. Each bar represents the mean±SEM values for experimental replicates. **p < 0.01 compared to A/Y siRNA/HA-construct (one-tailed). (D) Reconstitution of the β-complex in AUF-1-rescued or YB-1-rescued K562 cells. Top: R-EMSA analyses were conducted on a [32P]-labeled RNA corresponding to the β-globin 3′UTR, using cytoplasmic extracts from AUF-1-depleted and/or YB-1-depleted cells, or from similar cells expressing siRNA-resistant HA-AUF-1 or HA-YB-1. Arrow indicates the β-complex. Bottom: Western-transfer analyses of the same extracts, using a β-actin antibody.
2.4. β-complex assembly is enhanced by an mRNA poly(A) tail
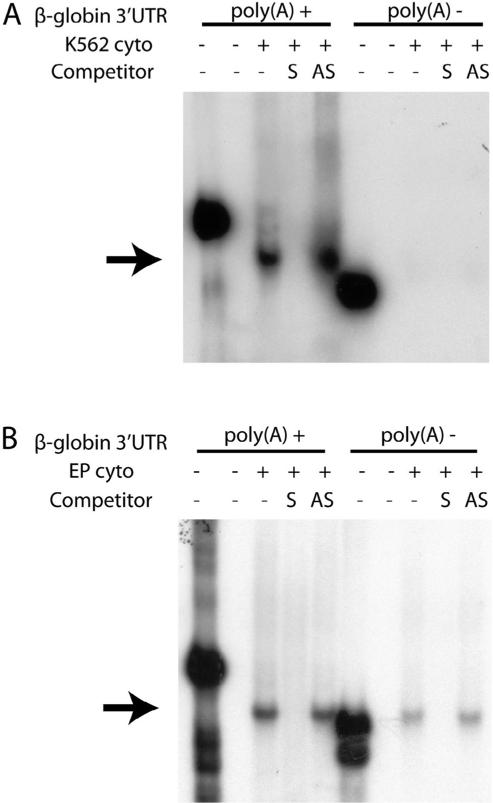
mRNA-stabilizing factors that target the 3′UTR characteristically act by enhancing the interaction between cytoplasmic poly(A) binding protein (PABPC1) and the polyadenylate tail (Kedar et al., 2010; Nagaoka et al., 2006; Wang et al., 1999). As both AUF-1 and YB-1 interact with PABPC1 when bound to heterologous mRNAs (Higashi et al., 2011; Laroia et al., 1999), we posited that the efficiency of β-complex assembly would be enhanced on β-globin 3′UTRs that contained a 3′ poly(A) sequence. This expectation was validated by EMSA analyses, conducted in K562 cells, that demonstrated the assembly of a strong β-complex on a polyadenylated β-globin 3′UTR, but not on a control non-adenylated β-globin 3′UTR (Fig. 4A). Parallel R-EMSAs conducted with extracts from primary human erythroid progenitor cells displayed a similar though less-pronounced difference that confirmed the principle of this effect (Fig. 4B). Collectively, these experiments demonstrate a structural interaction and implicate a functional relationship between the β-complex and the β-globin mRNA poly(A) tail.
Fig. 4.
A poly(A) tail enhances β-complex assembly on the β-globin 3′UTR. (A) Assembly of the β-complex is enhanced on a polyadenylated β-globin 3′UTR. R-EMSA analyses were conducted on β-globin 3′UTR RNAs that either did (+) or did not (−) contain an 18-nt poly(A) tail, using cytoplasmic extract prepared from K562 cells. Sense (S) and antisense (AS) DNA competitors verify the identity of the β-complex (arrow); 20% of each undigested probe was loaded as a control for transcript intensity and integrity. (B) Polyadenylation enhances β-complex assembly on β-globin mRNA 3′UTRs in primary human erythroid cells. As in panel A, using cytoplasmic extract purified from d10 EPO-induced CD34+ erythroid progenitor cells.
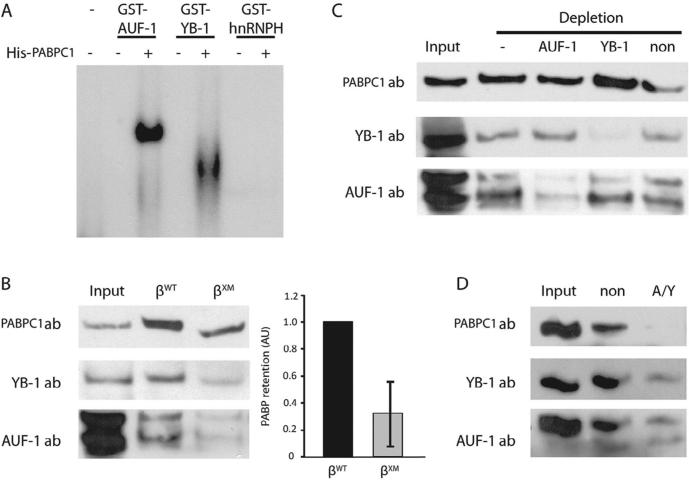
2.5. β-complex assembly is enhanced by interaction between PABPC1 and either AUF-1 or YB-1
The efficient assembly of the β-complex in the presence of a 3′ polyadenylate tail suggested that AUF-1 and YB-1 interact with PABPC1 to enhance protection of the β-globin poly(A) tail from constitutive degradative processes. We employed two independent strategies to investigate this hypothesis. First, we tested whether PABPC1 contributes to the efficiency with which AUF-1 and YB-1 bind to the β-globin 3′UTR, using an R-EMSA strategy in which a β-globin mRNA probe was incubated with the epitope-tagged factors, either alone or in the presence of PABPC1. Critically, both AUF-1 and YB-1 independently assemble an mRNP on the β-globin 3′UTR in the presence of PABPC1, but not in its absence (Fig. 5A), demonstrating a previously unknown requirement for PABPC1 in β-complex assembly. Second, we investigated the reciprocal possibility that the β-complex enhances the affinity of PABPC1 for the poly(A) tail, using an RNA affinity-capture strategy in which agaroseimmobilized, polyadenylated β-globin 3′UTR RNAs were incubated with K562 cytoplasmic extract. In contrast to the βWT 3′UTR, the βXM 3′UTR – which does not fully retain AUF-1 or YB-1 – demonstrated an ~70% decrease in retained PABPC1 (Fig. 5B), associating the efficiency of YB-1/AUF-1 binding to the integrity of the PABPC1-poly(A) tail interaction.
Fig. 5.
3′UTR-bound AUF-1 and YB-1 independently interact with PABPC1. (A) PABPC1 supports assembly of β-globin 3′UTR mRNPs that contain either AUF-1 or YB-1. R-EMSAs were conducted on a [32P]-labeled β-globin 3′UTR using GST-AUF-1, GST-YB-1, or negative control GST-hnRNP H (an RBP that does not form an mRNP) (Russo et al., 2010), and control GST. Reactions were amended with His-PABPC1 where indicated. (B) Ablation of the β-complex decreases PABPC1 retention. K562 cytoplasmic extracts were incubated with agarose-immobilized βWT and βXM 3′UTR RNAs. Retained proteins were resolved by SDS–PAGE and probed for PABPC1, YB-1, and AUF-1. Input extract was loaded at 5%. Left: Representative Western blots. Right: Quantification of retained PABPC1. Bars represent the mean±SEM values for 5 experimental replicates; the quantity of PABPC1 retained on βWT RNA was arbitrarily assigned unit value. (C) β-Globin 3′UTR-bound AUF-1 and YB-1 independently retain PABPC1. As in panel B, but K562 cytoplasmic extracts were initially depleted of either AUF-1 or YB-1 by repression of a corresponding shRNA, and subsequently incubated with an agarose-immobilized βWT 3′UTR RNA. Extracts from non-transfected (−) and non-coding shRNA-transfected cells (non) were used as negative controls. Results are representative of three independent replicates. (D) β-Globin mRNA does not retain PABPC1 from extracts that have been depleted of both AUF-1 and YB-1. As in panel B, but agarose-immobilized βWT 3′UTR RNAs were incubated with K562 cytoplasmic extracts that were depleted of both AUF-1 and YB-1 by expression of the corresponding shRNA. Extract from non-coding shRNA-transfected K562 cells was used as a negative control. Results are representative of three independent replicates.
As AUF-1 and YB-1 both assemble independent mRNPs on the β-globin 3′UTR (Fig. 3D), and exert redundant functions (Fig. 3C), we predicted that each factor alone would also be sufficient to enhance PABPC1 binding. We investigated this possibility in RNA affinity-capture experiments in which an agarose-immobilized βWT 3′UTR probe (Sup Fig. S3) was incubated in K562 extract that has previously been depleted of either AUF-1 or YB-1. PABPC1 was retained to a similar extent in the absence of either AUF-1 or YB-1 (Fig. 5C), but did not bind when AUF-1 and YB-1 were coordinately depleted (Fig. 5D), confirming the hypothesis that AUF-1 and YB-1 independently enhance PABPC1 binding to the poly(A) tail, and providing direct evidence for a mechanism through which the β-complex regulates the cytoplasmic stability of β-globin mRNA.
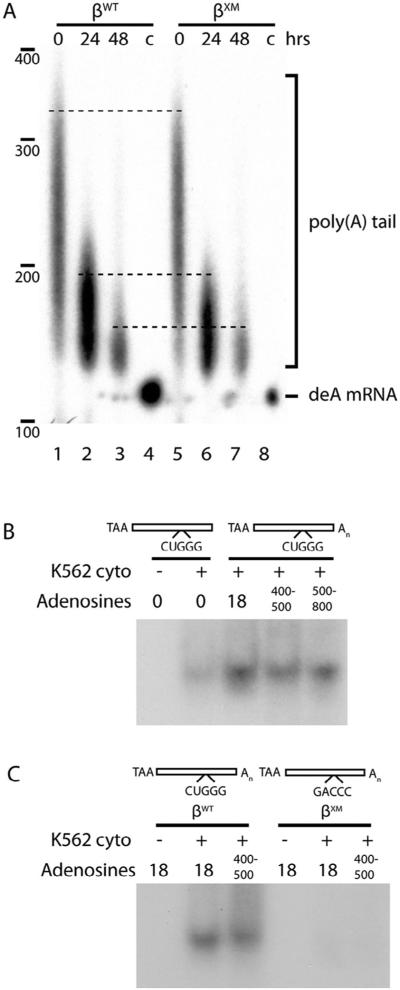
2.6. The β-complex protects the proximal β-globin mRNA poly(A) tail from deadenylation
The poly(A) tail-dependent interactions that we demonstrate suggest a mechanism through which the β-complex enhances β-globin mRNA stability by facilitating the poly(A)-protective properties of PABPC1. This model is consistent with our observations that the β-complex assembles most efficiently on adenylated RNAs (Fig. 4), interacts with PABPC1 (Fig. 5), and increases the half-life of β-globin mRNA (Fig. 2).This model also accords with the general mechanism for mRNA decay that initiates with 3′→5′ deadenylation, but does not proceed to 5′-decapping and subsequent endonucleolytic steps until the 5′-most region of the poly(A) tail is removed (Parker and Song, 2004; Wahle and Winkler, 2013). We characterized the poly(A) tails of stable βWT or unstable βXM mRNAs in K562 cells, and observed that their lengths were equivalent both at steady state (Fig. 6A, lanes 1 and 5), and following transcriptional arrest (lanes 2–3, 6–7). This result indicates that while the β-complex is critical to establishing or maintaining PABPC1-poly(A) structural interactions involving the initial 18 adenylate residues (Figs. 4 and 5), it has relatively little impact on PABPC1-poly(A) tail interactions beyond this point (Fig. 6A). R-EMSA analyses validated this possibility, demonstrating that β-complex assembly – although requiring a poly(A) tail – is not additionally enhanced by its elongation beyond the initial ~20 residues (Fig. 6B). Importantly, an elongated poly(A) tail did not facilitate β-complex assembly on the βXM 3′UTR (Fig. 6C), consistent with results from earlier studies conducted with minimally polyadenylated 3′UTRs. These analyses confirm that the β-complex is critical to maintaining a protective interaction between the poly(A) tail and PABPC1, thereby inhibiting the initial step of the mRNA-decay process.
Fig. 6.
The β-complex promotes binding of PABPC1 to the proximal region of the poly(A) tail. (A) Abolition of the β-complex does not materially affect the length of the β-globin mRNA poly(A) tail. Stably expressed βWT and βXM mRNAs were recovered from K562 cells at defined intervals following transcriptional arrest with tetracycline, and poly(A) tail lengths determined by ligase-mediated RT-PCR. The migration of deadenylated β-globin mRNA (deA mRNA; lanes 4, 8) was used as negative control. (B) The efficiency of β-complex assembly requires a minimal poly(A) tail, but is not enhanced by additional polyadenylate sequence. R-EMSAs were conducted on a [32P]-labeled β-globin 3′UTR RNAs containing poly(A) tails of varying lengths (Sup Fig. S4A), using cytoplasmic extracts from K562 cells. (C) The β-complex does not assemble on βXM RNA that contains an elongated poly(A) tail. As in panel B, using [ 32P]-labeled βWT and βXM 3′UTR RNAs containing variably-lengthed poly(A) tails (Sup Fig. S4B).
3. Discussion
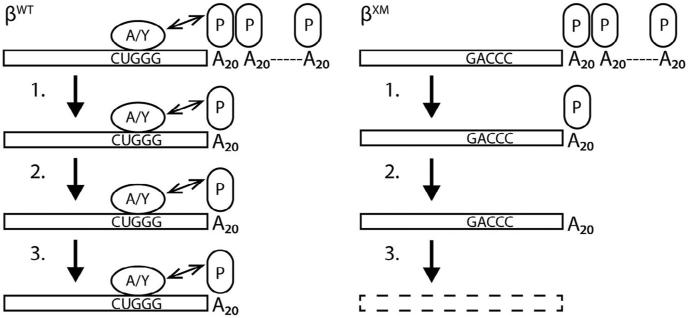
Although β-globin mRNA is among the most stable transcripts expressed in mammalian cells, the mechanism responsible for its unusually long half-life remains poorly understood. We recently demonstrated that two RNA-binding proteins, AUF-1 and YB-1, increase cytoplasmic levels of β-globin mRNA in erythroid cells by assembling an mRNP β-complex on its 3′UTR (van Zalen et al., 2012). The positioning of the β-complex as well as the mRNA stability-enhancing characteristics of its components combine to suggest that the β-complex functions by prolonging the half-life of β-globin mRNA. Here, we demonstrate that erythroid progenitors accumulate both β-globin mRNA and its encoded protein at the same stage of differentiation when they first support β-complex assembly. Our analyses also indicate that the β-complex prolongs the half-life of β-globin mRNA by strengthening the interaction between PABPC1 and the 5′-most end of the poly(A) tail, a function that either AUF-1 or YB-1 can perform independently (Fig. 7). These observations are fully consistent with the cell biology of erythropoiesis, where the β-complex is critical to post-transcriptional regulation of β-globin mRNA during late stages of terminal differentiation when erythroid progenitors are transcriptionally silent.
Fig. 7.
Model for β-complex-facilitated inhibition of cytoplasmic β-globin mRNA decay. The poly(A) tails of mRNAs that do (βWT) or do not (βXM) assemble a β-complex are initially of equal length and are similarly protected by polymeric PABPC1 (P) in erythroid cells. (1) During terminal erythroid differentiation, the poly(A) tails of both mRNAs shorten at the same rate, until a single PABPC1 monomer remains. (2) The binding of the 5′-most PABPC1 monomer to the poly(A) tail is enhanced by an interaction with either AUF-1 (A) or YB-1 (Y). (3) The enhanced interaction between the 5′-most PABPC1 monomer and the poly(A) tail protects the βWT mRNA from mRNA-decay processes that accelerate degradation of βXM and other variant β-globin mRNAs that do not assemble a β-complex. The double-headed arrow linking the β-complex to PABPC1 indicates a functional interaction; the structural relationship between these two elements has not yet been determined.
Our structural and functional analyses are consistent in demonstrating that AUF-1 and YB-1 act redundantly in erythroid cells: both proteins can independently assemble a β-globin mRNP (Figs. 3D, 5A), recruit PABPC1 (Fig. 5), and regulate levels of β-globin mRNA (Fig. 3C). While functional redundancy is not uncommon in processes that control human gene expression, this arrangement typically involves regulatory proteins that are structurally related (Melko et al., 2011; Paik et al., 2007). This is not the case for AUF-1 and YB-1, which belong to the heterogeneous nuclear ribonucleoprotein- and cold-shock protein families, respectively, suggesting a highly unusual basis for redundancy whose biological explanation remains to be established (Pinol-Roma et al., 1988; Wolffe et al., 1992). We have not, for example, excluded the possibility that AUF-1 and YB-1 exhibit distinct temporal patterns of expression during erythroid differentiation: while levels of both factors initially increase in EPO-induced CD34+ cells, our analyses did not examine the expression of either RBP at later, post-nuclear stages of differentiation (van Zalen et al., 2012). We have also considered that AUF-1/YB-1 functional redundancy might comprise a biological ‘fail-safe’ strategy for stabilizing the β-globin mRNA at a critical stage in erythroid differentiation. This potential cellular mechanism is supported by precedent: gld-1 mRNA, a factor that is essential for oogenic meiosis in Caenorhabditis elegans differentiation (Francis et al., 1995), is activated by two related poly(A) polymerases that converge on its 3′UTR to independently repress the action of the deadenylase CCR-4, maintaining poly(A) tail length and allowing for normal differentiation (Schmid et al., 2009). Considering the critical importance of β-globin mRNA to normal hemoglobinization, such a ‘fail-safe’ mechanism might not be entirely unanticipated in erythroid differentiation.
The expectation that AUF-1 and YB-1 act to stabilize β-globin mRNA in erythroid cytoplasm was originally based upon experimentally discrete observations that were fully consistent with this possibility. Both proteins bind to the β-globin 3′UTR in erythroid cytoplasm (van Zalen et al., 2012), and both can independently stabilize heterologous mRNAs (Capowski et al., 2001; Fellows et al., 2012; McGray et al., 2011). Moreover, AUF-1 and YB-1 both interact with PABPC1 that is bound to heterologous mRNAs (Higashi et al., 2011; Laroia et al., 1999), ostensibly protecting their poly(A) tails from decay rate-limiting deadenylation (Parker and Song, 2004; Wahle and Winkler, 2013). This last activity may be germane to the regulation of β-globin mRNA, as deadenylation appears to be the rate-limiting step for its decay as well (Gorski et al., 1975; Xu et al., 1998). Our analyses, which demonstrate that the degradation of β-globin mRNA is accompanied by a parallel shortening of the poly(A) tail (Fig. 6A), seem to confirm this expectation. The fact that the β-complex has relatively little effect on poly(A) tail length, but cannot assemble in the absence of a minimal poly(A) tail, suggests that the mRNP acts at a critical point in the β-globin mRNA decay process; specifically, at the time when the final PABPC1 monomer is removed from the proximal stretch of poly(A) tail. This is the step that irreversibly initiates mRNA decay (Parker and Song, 2004; Wahle and Winkler, 2013) and is thus a critical control point in regulating mRNA stability.
The importance of mRNA stability in cells that are transcriptionally silenced cannot be overstated. A 30% reduction in the cytoplasmic half-life of a specific mRNA predicts a corresponding deficit in its cytoplasmic level in transcriptionally active cells, but significantly overestimates its cytoplasmic level in transcriptionally silent cells. A consequence of this effect is that the relative quantities of two mRNAs with different t1/2 values will increase exponentially over time in transcriptionally silent cells, predicting that even a modest difference in the half-lives of βWT and βXM mRNAs would produce a significant discrepancy in their levels during the 3–5 day interval of transcriptional silence that characterizes terminal erythroid differentiation (Greer et al., 2009). We calculate that the ~4-hr increase in t1/2 value that we observe for βWT mRNA would allow for a 5- to 16-fold enrichment in its overall level during this developmental interval. The biological importance of this process is illustrated by common, natural occurring mutations that shorten the t1/2 value of human α-globin mRNA, effecting a 5-fold reduction in its accumulation within 3 days of transcriptional silencing (Morales et al., 1997; Weiss and Liebhaber, 1994). Similar regulatory defects that reduce the level of β-globin mRNA would be predicted to lead to a significant discordance in α- and β-globin chain synthesis that defines congenital genetic conditions that include the β thalassemias (Chaisue et al., 2007; Ranjbaran et al., 2013).
In addition to their mRNA-stabilizing properties, both AUF-1 and YB-1 interact with heterologous mRNAs to effect mRNA splicing and enhance translational efficiency. For example,YB-1 participates in splicing reactions, both as a component of the spliceosome during A- and B-complex formation, and as a co-factor in complexes that stimulate alternative splicing of specific pre-mRNAs (Wei et al., 2012). 3′UTR-bound YB-1 also selectively inhibits translation of several mRNAs (Lyabin et al., 2011; Paranjape and Harris, 2007), while AUF-1 promotes the translation of mRNAs encoding TAK1 and MYC (Liao et al., 2007; Sarkar et al., 2011).The observed structural interaction between PABPC1 and either AUF-1 or YB-1 suggests that the β-complex additionally influences β-globin mRNA translation, since PABPC1 is described to interact with initiation factors eIF4G and eIF4E to form a ‘closed loop’ structure of mRNA that may promote translation initiation and termination (Tarun and Sachs, 1996; Wells et al., 1998). We found no evidence to support a role for AUF-1 and YB-1 in either the splicing or translation of β-globin mRNA (data not shown), implicating their principal functions as mRNA-stabilizing factors for this transcript. Several other RBPs that interact with the β-globin 3′UTR – two in the nucleus (Jiang et al., 2006; Millevoi et al., 2009) – might mediate processes that regulate β-globin pre-mRNA in the nucleus before the mature transcript is exported to the cytoplasm.
Several observations suggest that AUF-1 and YB-1 – in addition to regulating β-globin mRNA stability – play a role in post-transcriptional mechanisms that direct terminal erythroid differentiation. Among other defects, a murine YB-1 loss-of-function model exhibits a severe failure of erythroid development; moreover, expression of YB-1 is regulated by GATA-1, a master regulator of erythroid development (Yokoyama, Harigae, Takahasi, Kameoka et al., 2003; Yokoyama, Harigae, Takahashi, Takahasi et al., 2003). Likewise, changes in AUF-1 in vivo levels can affect the expression of cyclin D1 and other factors that regulate normal erythropoiesis (Gouble et al., 2002; Lu et al., 2006). Finally, both YB-1 and AUF-1 regulate the stabilities of various mRNAs encoding proteins that are involved in erythroid cell growth and differentiation during erythropoiesis, such as GM-CSF, p16, and thymidylate synthase (Capowski et al., 2001; Pullmann et al., 2006; Wang et al., 2005). This evidence suggests that YB-1 and AUF-1 participate in an mRNA stability regulatory program that affects a larger number of mRNAs that are essential for normal terminal erythroid differentiation. It is also reasonable to speculate that these and other non-globin mRNAs that assemble mRNPs from AUF1 and YB1 in non-hematopoietic cells may be regulated through similar effects on their half-lives.
4. Experimental procedures
4.1. Plasmids
pTRE-β-WT was constructed by cloning a 3.0-kb fragment of human genomic DNA, encompassing the 1.5-kb full-length β-globin gene and its contiguous 3′-flanking region, into the SacII-EcoRV restriction sites of pTRE-Tight-BI-AcGFP1 (Clontech). A variant β-globin gene (βXM), containing a 3′UTR site-specific CTGGG→GACCC mutation at 3′UTR positions 65–69 (Sup Fig. S1), was generated by exchanging the 0.2-kb BstXI-SwaI fragment of pTRE-β-WT (encompassing the β-globin 3′UTR) with a corresponding synthetic DNA (Genscript) (van Zalen et al., 2012). Templates for in vitro transcription of human β-globin 3′UTRs, either without or with a 18-nt poly(A) tail, were constructed by directional cloning of the cognate DNA into the XhoI-BglII restriction sites of pSP72 (Promega). Plasmids encoding hem-agglutinin (HA)-tagged proteins were constructed by cloning AUF-1 (RefSeq NM_002138.3) or YB-1 (NM_004559.3) open-reading-frame DNAs (Genscript) into the unique NheI-NotI restriction sites of pTRE2pur-HA (Clontech), downstream and in-frame to the HA-encoding sequence. The AUF-1 and YB-1 coding regions were synthesized to contain synonymous codons that prevent base-paring with factor-specific siRNAs, but do not alter the native amino-acid sequences (described below). pTRIPz-YB-1, which encodes an shRNA that targets YB-1 mRNA, was purchased from Open Biosystems. Sequences encompassing AUF-1 and non-silencing specific shRNAs (Genscript) were exchanged for the YB-1 shRNA sequence using the unique MluIXhoI restriction sites. Plasmids encoding a luciferase ORF were modified to include β-globin 3′UTRs and 1.5 kb of contiguous 3′-flanking region by cloning the corresponding PCR-generated products into the unique XbaI-BamHI restriction sites of pGL3-Promoter (Promega). All recombinant sequences were verified by automated dideoxy sequencing.
4.2. RNA electromobility shift assays (R-EMSAs)
Template DNAs were linearized with BglII, and [α-32P]-labeled RNAs transcribed in vitro using SP6 polymerase under conditions recommended by the manufacturer (Ambion). To generate probes with elongated poly(A) tails,[32P]-labeled β-globin 3′UTRs containing a standard 18-nt poly(A) tail were incubated for 10 min at 37°C with either 1 U or 4 U of Escherichia coli poly(A) polymerase (NEB). The structural integrities of all RNAs were verified by denaturing acrylamide-urea gel electrophoreses.Transcripts (3 × 104 cpm) were incubated at RT for 30 min in EMSA buffer (10 mM Tris pH 7.4, 150 mM KCl, 1.5 mM MgCl2, 1 mM DTT) and supplemented with either 10 μg cellular extract or 250 ng recombinant protein. GST-AUF-1, GST-YB-1, and GST-hnRNP H (RefSeq NM_001257293.1) were purified as described (van Zalen et al., 2012), while His-PABPC1 (NM_002568) was commercially sourced (Abcam). Reaction products were digested at RT for 10 min with 20 U RNase T1 (Roche), resolved on a native 5% polyacrylamide gel, and exposed to X-ray film. Competitor DNAs (0.2 nmol) were added when indicated.
4.3. Cell culture
K562 and HEK293 cells were maintained at 37°C in a humidified 5% CO2 incubator in RPMI 1640 + GlutaMAX and DMEM media, respectively, and amended with 10% Tet System Approved FBS (Clontech) and 1× Antibiotic-Antimycotic (Invitrogen). K562/tTA cell lines were generated by transfecting K562 cells with pTet-Off Advanced (Clontech) and selecting for G418-resistant clones (400 μg/mL). Tetracycline (tet) responsiveness was assessed by transiently co-transfecting individual K562/tTA clones with pTRE-Tight-Luc (firefly luciferase) and transfectionefficiency control pRL (renilla luciferase), exposing cells to tet (1.0 μg/mL) for 48 hr, and quantifying the change in the firefly:renilla luciferase ratio relative to that in control tetnaïve cells. A single K562/tTA clone with the strongest tet response was used for all subsequent experiments. K562/tTA-β cells, which stably express high levels of either βWT or βXM mRNAs, were generated from K562/tTA cells by Amaxa electroporation (Lonza) with either pTRE-Tight-βWT or -βXM, and selected for resistance to hygromycin B (400 μg/mL).
4.4. K562 cell transfection
K562/tTA-β cells were subcultured for two days to a density of 0.5–1.0 × 106/mL, and 2–5 × 106 cells subsequently coelectroporated with 50 pmol of siRNA and 3.0 μg of HA-vector. After a 48-hr recovery, cells were aliquoted for RNA or cytoplasmic protein extraction. siRNAs specific for AUF-1, YB-1, and lamin A/C were purchased from Dharmacon.
4.5. Human primary cell culture
Human CD34+ cells were isolated, cultured, and differentiated as we previously describe (van Zalen et al., 2012). In brief, fresh umbilical cord blood was obtained from the University of Pennsylvania Stem Cell and Xenograft Core under an existing Institutional Review Board-approved protocol. Mononuclear cells were isolated on a Ficoll-Paque PLUS gradient (GE Healthcare) and immunomagnetically enriched for CD34+ cells according to the manufacturer's protocol (MiltenyiBiotec). Cells were cultured in IMDM (Invitrogen) containing 20% BIT 9500 Serum Substitute (Stem Cell Technologies) and 1× Antibiotic-Antimycotic, and induced to erythroid differentiation as described (van Zalen et al., 2012). Flow cytometric analyses were conducted with APC-CD45, PE-GlyA, and FITC-CD71.
4.6. Lentiviral production and transduction of K562 cells
HEK293 cells (1 × 106) were pre-cultured in DMEM for 24 hr in a 60-mm dish, and medium refreshed two hr before transfection. pTRIPz (14.0 μg) was mixed with Trans-Lentiviral Packaging mix and Fugene, and layered over the pre-cultured HEK293 cells according to the manufacturers’ instructions (ThermoScientific; Roche). Lentiviral supernatants were harvested after 36 hr. K562 cells were overlayered with 900 μL medium containing 100 μL supernatant, 8 μg/mL polybrene, and 10 mM HEPES; centrifuged in 6-well plates at RT for 90 min at 3200 rpm; and selected for resistance to puromycin (2.0 μg/mL). shRNA expression was induced by adding 2.0 μg/mL doxycycline for 72 hr, and cells subsequently harvested for either RNA isolation or protein extraction.
4.7. Western transfer analyses
Cytoplasmic extracts were resolved on a pre-cast 4–12% gradient SDS–PAGE gel, and transferred to nitrocellulose using an XCell-II Blot Module (Invitrogen). Membranes were sequentially incubated for one hr at RT in blocking buffer (Superblock T20 PBS; ThermoScientific), and for one hr in blocking buffer supplemented with primary antibody. Membranes were washed with PBS+Tween-20 (0.05%) and incubated for one hr in blocking buffer containing horseradish peroxidase-conjugated secondary antibody (GE Healthcare).Washed membranes were analyzed by ECLPlus chemiluminescence as recommended by the manufacturer (GE Healthcare) and quantification performed using Image Lab 5.0 software (Biorad).Antibodies were purchased from Santa Cruz (β-globin, catalogue # sc-21757; PABPC1, sc-32318; YB-1, sc-101198; β-actin, sc-58673) or generously provided by G. Dreyfuss (University of Pennsylvania; AUF-1).
4.8. Ligation-mediated reverse-transcription PCR and poly(A)-tail mapping
An RNA oligonucleotide containing a polymerization-inhibiting deoxythymidine (idT) (5′-pAGUGAUUGACCUG ACGCAUAAGGAC(idT)-3′; Dharmacon) was ligated to 0.25– 1.0 μg total RNA for 3 hr at 37°C using T4 RNA ligase (NEB). The RNAs were reverse transcribed with RT primer (5′-CCTTATGCGTCAGGTCA-3′) for 60 min at 50°C using Superscript III RT (Invitrogen). First-strand cDNAs were then amplified using primers bracketing the poly(A) tail of the β-globin mRNA (reverse 5′-CCTTATGCGTCAGGTCAATCA-3′; forward 5′-TTCCTTT GTTCCCTAAGTCCAA-3′) for 25 cycles (94°Cx30s, 64°Cx30s, and 72°Cx60s). Amplicons were purified (QiaQuick, Qiagen), then re-amplified for 12 additional cycles in the presence of a nested [γ-32P]-5′-end-labeled forward primer (5′-TTGTTCCCTAAG TCCAACTACTAAA-3′); reaction products were then resolved on a 6%/8M polyacrylamide/urea gel. A deadenylated control was generated by digesting total RNA with RNase H (NEB) in the presence of excess oligo(dT)18 prior to ligation-mediated RT-PCR.
4.9. RT-qPCR
Total RNA was purified using Trizol and incubated with RNase-free DNase I (Roche). cDNAs were synthesized using Superscript III RT and quantitated by Taqman qPCR (Applied Biosystems) with FAM-MGM labeled probe sets (HBB, Applied Biosystems catalogue # Hs00747223_g1; YBX1, Hs00898625_g1; HNRNPD, Hs01086912_m1) and endogenous control VICMGM labeled β-actin (4352935) or GAPDH (4326317E) assays. Reactions were monitored on an SDS-7500 cycler (Applied Biosystems) and data analyzed with corresponding system software. Statistical significance was calculated by Student's t-test.
4.10. RNA capture affinity
Template DNAs were linearized with BglII, and RNAs transcribed in vitro in the presence of biotin-16-UTP (Ambion) using SP6 polymerase (Roche). Biotin-linked RNAs (5 μg) were incubated for one hr at 4°C with avidin-agarose beads (ThermoScientific). Pelleted beads were re-suspended with 200 μL K562 cytoplasmic extract in binding buffer (10 mM Tris pH 7.4, 150 mM KCl, 2.5 mM MgCl2, 0.04% glycerol, 1 mM DTT) and incubated for two hr at 4°C. The liganded beads were washed twice with PBS and twice with PBS+Triton X-100 (0.5%). Retained proteins were eluted in NuPAGE LDS Sample Buffer and resolved on a pre-cast 4–12% gradient SDS–PAGE gel (Invitrogen).
4.11. mRNA decay analyses
Two-point decay analyses were performed on duplicate aliquots of K562/tTA cells that were transiently co-transfected with pGL3 (Promega; 2.5 μg) and either pTRE-β-WT or pTRE-β-XM (2.5 μg) using Amaxa technology, and then cultured for 16 hr in the presence of doxycycline (0.2 μg/mL). Cells were washed twice with PBS and once with RPMI to permit a 4-hr pulse interval of β-globin gene expression, at which point dox (2.0 μg/mL) was restored to the culture medium. Cells were harvested 0 and 16 hr later withTrizol, and isolated RNAs subjected to RT-qPCR analyses (see above). Conventional half-life analyses were conducted using the reverse-chase strategy for estimating mRNA half-lives (Abdulmalik et al., 2012). 4 × 104 K562/tTA cells that had previously been engineered to stably express βWT or βXM genes were aliquoted in 180 μL media in a 96-well format, 24 hr prior to the start of the transcriptional chase. At t = 48 hr, cells were washed twice with excess PBS, then lysed in situ using Cells-to-Ct reagent (Ambion). Equal volumes of prepared RNAs were reverse-transcribed, and first-strand cDNAs quantified by RT-qPCR using Taqman assays. Levels of β-globin mRNAs were normalized to levels of control β-actin mRNAs, then regressed to an exponential decay function.
Supplementary Material
Acknowledgments
The authors thank G. Dreyfuss for sharing critical re-agents. This work was supported in part by a Research Fellowship from the Cooley's Anemia Foundation (SvZ) and by NIH grants HL082754 (JER) and HL61399 (JER).
Footnotes
Appendix: Supplementary material
Supplementary data to this article can be found online at doi:10.1016/j.mod.2015.02.003.
REFERENCES
- Abdulmalik O, Lombardi AA, Russell JE. A reverse time-course method for transcriptional chase analyses of mRNA half-lives in cultured cells. PLoS ONE. 2012;7:e40827. doi: 10.1371/journal.pone.0040827. doi:10.1371/journal.pone.0040827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J. Immunol. 2001;167:5970–5976. doi: 10.4049/jimmunol.167.10.5970. [DOI] [PubMed] [Google Scholar]
- Chaisue C, Kitcharoen S, Wilairat P, Jetsrisuparb A, Fucharoen G, Fucharoen S. Alpha/beta-Globin mRNA ratio determination by multiplex quantitative real-time reverse transcription-polymerase chain reaction as an indicator of globin gene function. Clin. Biochem. 2007;40:1373–1377. doi: 10.1016/j.clinbiochem.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, et al. Stability regulation of mRNA and the control of gene expression. Ann. N. Y. Acad. Sci. 2005;1058:196–204. doi: 10.1196/annals.1359.026. [DOI] [PubMed] [Google Scholar]
- Clegg JB, Weatherall DJ, Milner PF. Haemoglobin Constant Spring – a chain termination mutant? Nature. 1971;234:337–340. doi: 10.1038/234337a0. [DOI] [PubMed] [Google Scholar]
- Fellows A, Griffin ME, Petrella BL, Zhong L, Parvin-Nejad FP, Fava R, et al. AUF1/hnRNP D represses expression of VEGF in macrophages. Mol. Biol. Cell. 2012;23:1414–1422. doi: 10.1091/mbc.E11-06-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J, Morrison MR, Merkel CG, Lingrel JB. Poly(A) size class distribution in globin mRNAs as a function of time. Nature. 1975;253:749–751. doi: 10.1038/253749a0. [DOI] [PubMed] [Google Scholar]
- Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- Greer JP, Foerster J, Rodgers GM, Paraskevas F, Glader B, Arber AB, et al. Wintrobe's Clinical Hematology. 12th ed. Lippincott Williams & Wilkins; Philadelphia: 2009. [Google Scholar]
- Higashi K, Tomigahara Y, Shiraki H, Miyata K, Mikami T, Kimura T, et al. A novel small compound that promotes nuclear translocation of YB-1 ameliorates experimental hepatic fibrosis in mice. J. Biol. Chem. 2011;286:4485–4492. doi: 10.1074/jbc.M110.151936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, X., Xu S, Russell JE. A nucleolin-binding 3’ untranslated region element stabilizes beta-globin mRNA in vivo. Mol. Cell. Biol. 2006;26:2419–2429. doi: 10.1128/MCB.26.6.2419-2429.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedar VP, Darby MK, Williams JG, Blackshear PJ. Phosphorylation of human tristetraprolin in response to its interaction with the Cbl interacting protein CIN85. PLoS ONE. 2010;5:e9588. doi: 10.1371/journal.pone.0009588. doi:10.1371/journal.pone.0009588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyabin DN, I., Eliseeva A, Skabkina OV, Ovchinnikov LP. Interplay between Y-box-binding protein 1 (YB-1) and poly(A) binding protein (PABP) in specific regulation of YB-1 mRNA translation. RNA Biol. 2011;8:883–892. doi: 10.4161/rna.8.5.16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGray AJ, Gingerich T, Petrik JJ, Lamarre J. Regulation of thrombospondin-1 expression through AU-rich elements in the 3’UTR of the mRNA. Cell. Mol. Biol. Lett. 2011;16:55–68. doi: 10.2478/s11658-010-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melko M, Douguet D, Bensaid M, Zongaro S, Verheggen C, Gecz J, et al. Functional characterization of the AFF (AF4/FMR2) family of RNA-binding proteins: insights into the molecular pathology of FRAXE intellectual disability. Hum. Mol. Genet. 2011;20:1873–1885. doi: 10.1093/hmg/ddr069. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Decorsiere A, Loulergue C, Iacovoni J, Bernat S, Antoniou M, et al. A physical and functional link between splicing factors promotes pre-mRNA 3’ end processing. Nucleic Acids Res. 2009;37:4672–4683. doi: 10.1093/nar/gkp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes KC, Quaresma AJ, Maehnss K, Kobarg J. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D). Biol. Chem. 2003;384:25–37. doi: 10.1515/BC.2003.004. [DOI] [PubMed] [Google Scholar]
- Morales J, Russell JE, Liebhaber SA. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- Nagaoka K, Suzuki T, Kawano T, Imakawa K, Sakai S. Stability of casein mRNA is ensured by structural interactions between the 3’-untranslated region and poly(A) tail via the HuR and poly(A)-binding protein complex. Biochim. Biophys. Acta. 2006;1759:132–140. doi: 10.1016/j.bbaexp.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape SM, Harris E. Y box-binding protein-1 binds to the dengue virus 3’-untranslated region and mediates antiviral effects. J. Biol. Chem. 2007;282:30497–30508. doi: 10.1074/jbc.M705755200. [DOI] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Peixeiro I, Silva AL, Romao L. Control of human beta-globin mRNA stability and its impact on beta-thalassemia phenotype. Haematologica. 2011;96:905–913. doi: 10.3324/haematol.2010.039206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Pullmann R, Jr., Abdelmohsen K, Lal A, Martindale JL, Ladner RD, Gorospe M. Differential stability of thymidylate synthase 3’-untranslated region polymorphic variants regulated by AUF1. J. Biol. Chem. 2006;281:23456–23463. doi: 10.1074/jbc.M600282200. [DOI] [PubMed] [Google Scholar]
- Ranjbaran R, Okhovat MA, Mobarhanfard A, Aboualizadeh F, Abbasi M, Moezzi L, et al. Analysis of beta/alpha globin ratio by using relative qRT-PCR for diagnosis of beta-thalassemia carriers. J. Clin. Lab. Anal. 2013;27:267–271. doi: 10.1002/jcla.21594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Sullivan TD. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- Russo A, Siciliano G, Catillo M, Giangrande C, Amoresano A, Pucci P, et al. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim. Biophys. Acta. 2010;1799:419–428. doi: 10.1016/j.bbagrm.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Rutherford T, Clegg JB, Higgs DR, Jones RW, Thompson J, Weatherall DJ. Embryonic erythroid differentiation in the human leukemic cell line K562. Proc. Natl. Acad. Sci. U.S.A. 1981;78:348–352. doi: 10.1073/pnas.78.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, et al. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol. Cell. Biol. 2011;31:602–615. doi: 10.1128/MCB.00835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Kuchler B, Eckmann CR. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 2009;23:824–836. doi: 10.1101/gad.494009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3’-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr., Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- van Zalen S, Jeschke GR, Hexner EO, Russell JE. AUF-1 and YB-1 are critical determinants of beta-globin mRNA expression in erythroid cells. Blood. 2012;119:1045–1053. doi: 10.1182/blood-2011-10-387316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volloch V, Housman D. Stability of globin mRNA in terminally differentiating murine erythroleukemia cells. Cell. 1981;23:509–514. doi: 10.1016/0092-8674(81)90146-x. [DOI] [PubMed] [Google Scholar]
- Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-Not and Pan2-Pan3 complexes. Biochim. Biophys. Acta. 2013;1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Wang W, Martindale JL, Yang X, Chrest FJ, Gorospe M. Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 2005;6:158–164. doi: 10.1038/sj.embor.7400346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF, et al. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012;40:8622–8636. doi: 10.1093/nar/gks579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss IM, Liebhaber SA. Erythroid cell-specific determinants of alpha-globin mRNA stability. Mol. Cell. Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Gao M, Jones CL, Wilusz J, Peltz SW. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Wolffe AP, Tafuri S, Ranjan M, Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992;4:290–298. [PubMed] [Google Scholar]
- Xu N, Loflin P, Chen CY, Shyu AB. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 1998;26:558–565. doi: 10.1093/nar/26.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H, Harigae H, Takahashi S, Kameoka J, Miyamura K, Ishizawa K, et al. High expression of YB-1 gene in erythroid cells in patients with refractory anemia. Int. J. Hematol. 2003;78:213–218. doi: 10.1007/BF02983797. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Harigae H, Takahashi S, Takahashi S, Furuyama K, Kaku M, et al. Regulation of YB-1 gene expression by GATA transcription factors. Biochem. Biophys. Res. Commun. 2003;303:140–145. doi: 10.1016/s0006-291x(03)00296-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.