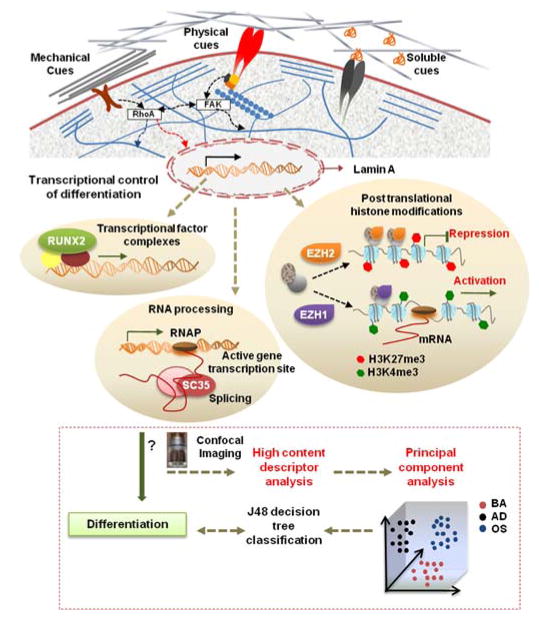
Fig. 1. Workflow highlighting utility of nuclear imaging-based profiling.
Cell-material interactions modulate nuclear protein organization through signaling cascades as well as cytoskeletal-nuclear links, which regulates nuclear programs such as activation of transcriptional factors, posttranslational histone modifications, and RNA processing that direct gene expression and cell fate. In this study, the influences of microenvironmental cues on organization of various sub-nuclear reporters involved in these nuclear programs, namely, Lamin A, SC-35, RUNX2, H3K4me3, and EZH2 was evaluated at early time-points during differentiation. hMSCs were cultured on different substrates in the presence of adipogenic (AD) or osteogenic (OS) or basal media (BA) for 3 days, and high resolution images were acquired using confocal microscopy. High content analysis was then done to compute Haralick texture descriptors that define the spatial organization of the nuclear reporter. This set of descriptors was then dimensionally reduced using Principal Component Analysis (PCA). Subsequently, the principal components were taken as an input for a classifier that used machine-learning approaches based on a J48 decision tree algorithm to classify cells exposed to different conditions. The predictive classification model was validated by correlations with 14-day endpoint assays. Using this framework, we can go through several iterations to analyze changes in nuclear reporter organization and thus screen and identify microenvironmental cues that elicit desired phenotypic responses.