Abstract
Lineage-specific stem cells are critical for the production and maintenance of specific cell types and tissues in multicellular organisms. In Arabidopsis, the initiation and proliferation of stomatal lineage cells is controlled by the basic helix-loop-helix transcription factor SPEECHLESS (SPCH). SPCH-driven asymmetric and self-renewing divisions allow flexibility in stomatal production and overall organ growth. How SPCH directs stomatal lineage cell behaviors, however, is unclear. Here, we improved the chromatin immunoprecipitation (ChIP) assay and profiled the genome-wide targets of Arabidopsis SPCH in vivo. We found that SPCH controls key regulators of cell fate and asymmetric cell divisions and modulates responsiveness to peptide and phytohormone-mediated intercellular communication. Our results delineate the molecular pathways that regulate an essential adult stem cell lineage in plants.
In multicellular organisms, the need to generate and maintain diverse cell types and tissues is fulfilled by lineage-specific stem cells (1). These stem cell lineages, active post-embryonically, produce a defined set of cell types. Although the origins of these lineage-specific stem cells during development are largely obscure, master transcription factors are implicated in their specification in both animals and plants (1–3). However, low expression levels and/or presence in limited number of cells makes genome-wide study of these transcriptional regulators by standard chromatin immunoprecipitation (ChIP) assays, the most common technique for studying protein-DNA interactions, technically challenging.
Stomata are epidermal valves that mediate gas exchange between the plant and atmosphere. In Arabidopsis, stomatal guard cells are derived from an epidermal cell lineage (Fig. 1A) (4, 5). Two populations of stomatal precursor stem cells, meristemoid mother cells and meristemoids, have limited self-renewing properties and proliferate without the benefit of a stem cell niche (4–6). These stem cells are created through the post-embryonic activity of SPEECHLESS (SPCH) in a subset of protodermal cells (7, 8). SPCH is a control point through which developmental, environmental and phytohormone signals are integrated (4, 5). However, no targets of SPCH have been reported and thus the sphere of its regulatory influence is unknown. Here we develop a ChIP method optimized for rare developmental regulators and profile the genome-wide binding of SPCH in vivo. In combination with multiple transcriptional response datasets, our ChIP-Seq data indicate that SPCH programs an entire lineage by promoting fate transitions and asymmetric cell divisions (ACDs). SPCH also modulates the sensitivity of stomatal lineage cells to hormone and peptide/receptor-mediated signaling. Our results suggest how this lineage exhibits significant autonomy while still coordinating with the overall organ development program.
Fig. 1. Chromatin immunoprecipitation (ChIP) optimized for cell-type-specific studies in vivo.
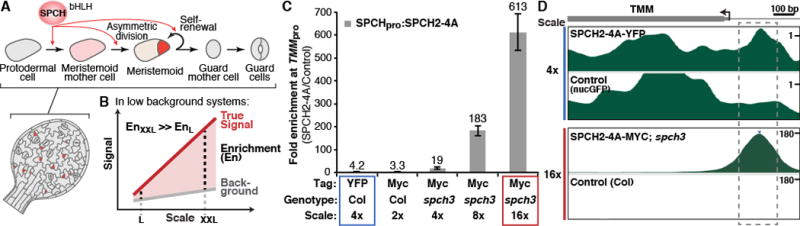
(A) Arabidopsis stomatal development scheme. SPCH controls the initiation and proliferation of the stem cell-like stomatal lineage precursors (pink and red cells). (B) Model for improving target enrichment in ChIPs through increasing experimental scale. (C and D) ChIPs at larger scales improve target enrichments. ChIP-qPCR assays of a SPCH variant on the TMM promoter performed at the indicated conditions (C). SPCH ChIP-Seq profiles at TMM (D) generated from ChIPs at 4× and 16× (blue and red box in C, respectively). The y-axis represents the enrichment values; note scales. Dashed box marks the SPCH-binding region.
Like many developmental regulators, SPCH expression is transient and limited to few cells (Fig. 1A). Standard ChIP assays on SPCH yielded only modest target enrichment (~4-fold, Fig. 1C, blue box) and thus we needed improved ChIP sensitivity for the detection of endogenously weak signals. We hypothesized that if background signals in a ChIP assay could be kept low, increasing the experimental scale would lead to a disproportional increase in signals from targets (true signal) over background (Fig. 1B). Therefore, performing ChIP at a large scale may achieve high target enrichment even for low abundance proteins. We tested this hypothesis with ChIPs at three different scales on a spch mutant line bearing a complementing, Myc-tagged SPCH variant driven by its native promoter (SPCHpro:SPCH2-4A-MYC; fig. S1). The scales represented 4, 8 and 16 times (or 6, 12 and 24 g) the input materials used in a typical Arabidopsis ChIP experiment. ChIP-qPCR assays of SPCH on the promoter of TOO MANY MOUTHS (TMM) showed that scale increase improves target enrichment up to 600-fold at 16x (or a >30-fold increase in enrichment with a 4-fold scale increase) (Fig. 1C, three rightmost columns). Thus, weak signals can be enhanced by maximizing input. We termed this method Maximized Objects for Better Enrichment (MOBE)-ChIP.
To profile genome-wide binding events of SPCH, we performed and pooled six MOBE-ChIPs on SPCHpro:SPCH2-4A-MYC and on a wild-type control for high-throughput sequencing (scale: 16x, total: 144 g/genotype; Fig. 1C, red box, and Fig. S2B). For comparison, standard ChIP-Seq was also included (pooled from 9 independent ChIPs on SPCHpro:SPCH2-4A-YFP and nucGFP at 4×; Fig. 1C, blue box, and fig. S2A). MOBE-ChIP-Seq confirmed the ChIP-qPCR results at the TMM promoter, revealing a single peak with an enrichment score of 178 (−log10(q-value): 1.2×106); the corresponding peak from our 4× run had a score of 1.2 (−log10(q-value): 5.7) (Fig. 1D). Low background signal is also a genome-wide trend. Using the peak-calling algorithm CSAR (9), we detected peaks with an enrichment score as low as 1.62 at a false discovery rate (FDR) of 1×10−6, in contrast to other studies whose peaks above threshold scores of 1.85 and 79.6 were detected at FDRs of 0.01 and 0.001, respectively (table S1) (10, 11). The ability to identify these low-coverage peaks is indicative of the power of signal enrichment. Thus, through MOBE-ChIP-Seq, we generated a comprehensive in vivo genome-wide binding map of SPCH.
Using two complementary peak-calling pipelines, we identified 8327 SPCH-bound regions (tables S2 and S3). 70% of the SPCH binding peaks are associated with gene promoters, mostly within 500 bp upstream of the transcriptional start site (Fig. 2A and fig. S3). De novo discovery of enriched motifs in the binding peaks identified CDCGTG as the top-scoring motif; this variant of the E-box (CACGTG), to which bHLH proteins typically bind, is enriched at the summit of the SPCH peaks (Fig. 2B and fig. S4).
Fig. 2. Genome-wide analysis of SPCH-binding targets reveals direct roles in lineage specification and asymmetric cell divisions.
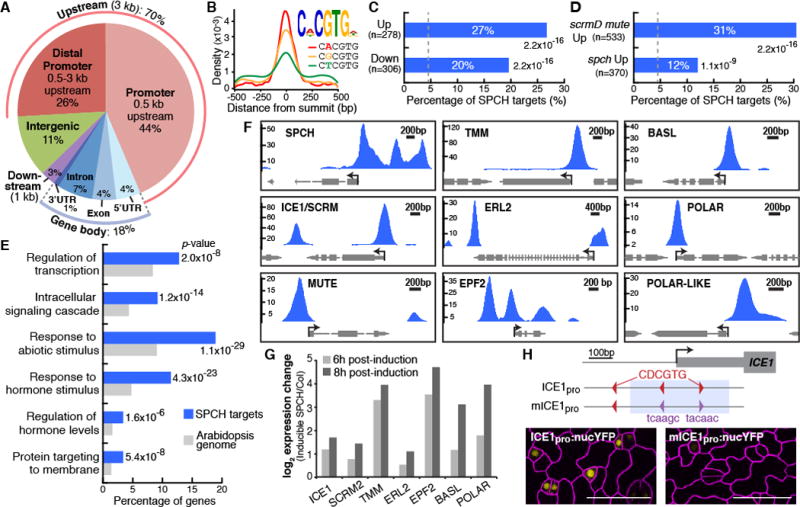
(A) Distribution of SPCH-binding peaks relative to gene structure. (B) Top-scoring motif (E-value: 7.5×10−365) and the position of its three variants in SPCH-binding peaks. (C and D) Percentage of SPCH targets among differentially-expressed genes in RNA-Seq analysis of inducible SPCH1-4A (C), and microarray analysis of meristemoid–enriched (scrmD mute) or –depleted (spch) mutants (13) (D). P-values are calculated by Fisher’s exact test. Dashed line indicates percentage by chance. (E) Select enriched GO terms of SPCH target genes. (F and G) SPCH binds and activates key stomatal regulators. ChIP-Seq profiles of select stomatal genes (F). The y-axis represents peak score (CSAR) and arrows indicate gene orientation and transcriptional start sites. Gene expression changes upon induction of SPCH in RNA-Seq analysis (G). (H) Importance of SPCH-binding motif (red) on ICE1 expression. Mutation of two motifs (purple; mICE1pro) within the SPCH-binding peak (blue shading) abrogates ICE1 expression (yellow). Confocal images of 4-dpg abaxial cotyledons have ML1pro:mCherry-RCI2A-marked cell outlines (purple). Scale bar, 40 μm.
To focus on loci most likely to respond transcriptionally to SPCH binding, we generated a “high-confidence” subset of peaks that were non-intergenic with enrichment scores ≥10 (table S2). Among the high-confidence targets, Gene Ontology (GO) terms for genes involved in regulation of transcription, signaling, response to stimulus and regulation of hormone levels are significantly enriched (Fig. 2E, fig. S5 and table S4). This suggests that in the initiation of the stomatal lineage, SPCH could act as a mediator of environmental and hormone inputs which are translated into further downstream transcriptional and signaling networks. The enrichment of the GO term, “protein targeting to membrane”, is interesting given the membrane-associated polarization of stomatal lineage proteins BASL and POLAR during asymmetric divisions (12, 13).
To correlate SPCH binding with transcriptional responses on a genome-wide scale, we compared the high-confidence SPCH targets to datasets representing genes expressed in response to SPCH induction (fig. S6 and table S5), and those enriched for genes preferentially expressed in the stomatal lineage (13) (fig. S7). Significant enrichment of the SPCH targets was found among genes both up- and down-regulated in response to SPCH induction (27 and 20%, respectively, Fig. 2C) and in plants with excess or no meristemoids (31 and 12%, Fig. 2D). By chance, SPCH would be predicted to bind to ~4.5% of genes in the datasets (1517 targets/33602 Arabidopsis genes). Overall, theses comparisons indicate that nearly a quarter (23%) of the SPCH targets are differentially expressed (table S6) and SPCH may activate or repress a large number of its targets directly.
Meristemoid-active stomatal regulators are among the direct SPCH targets (Fig. 2F, fig. S8A, fig. S9 and table S7). SPCH binds to its own promoter and to the promoter of its heterodimeric bHLH partners, ICE1/SCRM and SCRM2 (14), and induces their expression (Fig. 2, F and G, and fig. S8A). Although initial activation of SPCH may not require SPCH protein (fig. S10), this positive feedback loop may be an essential part of a bistable switch that converts the initially low and stochastic expression of SPCH into an active SPCH-SCRM heterodimer to drive stomatal lineage fates. SPCH also binds and activates expression of genes encoding the secreted ligand EPF2, the receptor TMM and the ERECTA family of receptor-like kinases (Fig. 2, F and G, and fig. S8A), all of which enforce proper patterning by restricting proliferation in the early stomatal lineage and act upstream of kinases that target SPCH for posttranslational down-regulation (4, 15–17). Further, SPCH binds to the promoters and activates expression of polarly-localized proteins, BASL and POLAR, suggesting a direct role in regulating the ACD process (Fig. 2, F and G). SPCH binding is not associated with a later expressed stomatal lineage EPF (EPF1), with EPFs not expressed in the stomatal lineage (CHALLAH) or with the broadly expressed MAPKKK YODA (fig. S8B). Taken together, our ChIP-Seq and RNA-Seq data reveal the broad and direct roles of SPCH in sustaining a SPCH transcriptional cascade, establishing meristemoid identity and mediating ACDs.
The CDCGTG motif appears in the SPCH-bound regions of stomatal targets like ICE1, TMM and ERL2. In ICE1, SPCH binds in two peaks centered on the locations of two CDCGTG motifs (fig. S11). To test the role of SPCH-binding motifs in ICE1 expression, we generated a reporter bearing point mutations in the two peak-associated motifs (Fig. 2H) to compare to the WT reporter. Consistent with previous reports (14), expression of the WT promoter reporter (ICE1pro) was observed in the stomatal lineage; however, the mutant reporter (mICE1pro) was nearly undetectable (Fig. 2H and fig. S12). Similar dependence was seen with SPCH-up-regulated gene At2g34510, which contains CDCGTG within a strong intronic SPCH binding peak. Deletion of the SPCH binding region abrogated early stomatal lineage-specific expression (Fig. S13).
An intriguing meristemoid behavior is the ability to self-renew through ACDs. Beyond requirements for SPCH activity and the polarly-localized BASL (Fig. 2F), however, little is known about the ACD process. Among SPCH targets, ARK3/AtKINUa (Fig. 3A) caught our attention as a plant-specific kinesin in the preprophase band (18). In plants, the preprophase band marks the future division plane (19). Confocal analysis of ARK3pro:ARK3-YFP showed localization to preprophase bands of asymmetrically-dividing meristemoids (Fig. 3B). Co-expression with SPCHpro:SPCH-CFP indicated that SPCH precedes ARK3, consistent with SPCH activating ARK3 expression (Fig. 3, C to E). To ascertain its function in the stomatal lineage, we reduced ARK3 expression by driving an artificial microRNA against it with the SPCH promoter (SPCHpro:amiR-ark3). In the cotyledon epidermis of amiR-ark3 expressing plants, we observed clusters of meristemoid-like small cells at 4 days post-germination that developed into clusters of stomata at 11 days (Fig. 3, G and I, brackets). These small cell clusters, which displayed diminished physical asymmetry, appear to arise from misplaced but complete division planes. Significantly, cell wall stubs or other evidence of incomplete divisions were not observed. The amiR-ark3 phenotypes resembled those associated with basl mutants (12) and are hallmarks of loss of ACD capacity. Thus, ARK3 appears to be a new player essential for ACD, possibly through regulating preprophase band placement, and establishes a direct link between SPCH and the ACD machinery.
Fig. 3. SPCH regulates asymmetric cell division (ACD) through a preprophase band-localized kinesin.
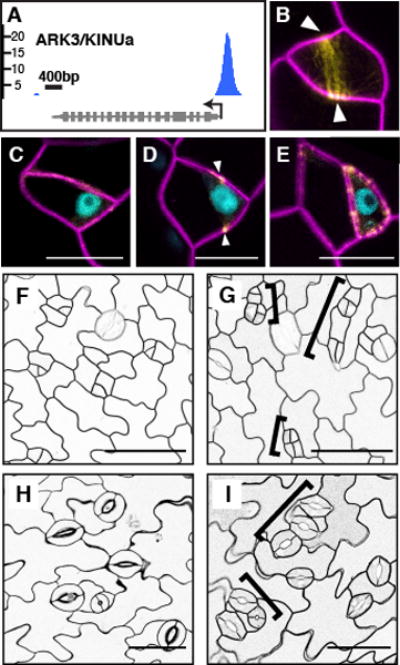
(A) SPCH ChIP-Seq profile of ARK3/KINUa. (B to E) Expression of ARK3pro:ARK3-YFP (yellow) and its co-expression with SPCHpro:SPCH-CFP (blue) (C to E only) before (C), during (B and D) and after (E) a stomatal ACD. Arrowheads indicate the preprophase band. (F to I) ACD defects in SPCHpro:amiR-ark3 (G and I), compared to Col (F and H). Brackets mark clusters of small cells (G) or guard cells (I). Confocal images are of 3- (B to E), 4- (F and G) and 11-day (H and I) abaxial cotyledons with ML1pro:mCherry-RCI2A-marked cell outlines. Scale bars, 10 μm (C to E), 50 μm (F to I).
SPCH initiates a lineage with autonomous control over cell division and fate determination. Nonetheless, the stomatal lineage is also coordinated with developmental programs operating across tissues and organs. Phytohormones play critical roles in coordinating development and recent reports indicate auxin, brassinosteroid (BR) and abscisic acid regulate stomatal development (20–23). BR controls stomatal development through phosphorylation of YODA and SPCH by its central GSK3-like kinase, BIN2 (Fig. 4F) (21, 22). Among SPCH target categories, BR biosynthetic and response genes show significant enrichment (fig. S14). Notably, SPCH binds to the promoters of BIN2 and CPD, an essential enzyme for BR biosynthesis (Fig. 4, A and F) and absence of CPD results in stomatal overproduction (24). We tested the effect of SPCH on the expression of BR genes by RT-qPCR in the meristemoid-enriched line SPCHpro: SPCH2-4A-YFP (Fig. 4B). Consistent with inhibition of BR signaling, we found that BIN2 expression is elevated, whereas CPD is repressed (Fig. 4B). Supporting BIN2’s role in promoting SPCH function, stomatal lineage-specific expression of BIN2-1 led to small cell clusters in cotyledons, similar to those observed upon SPCH overexpression (Fig. 4C). Thus, our results suggest the presence of feedback by SPCH counteracting BR signaling (Fig. 4F). SPCH also binds to the BR signaling effectors, the BZR1 family of transcription factors (BZR1, BES1/BZR2, BEH1 to 4), and BIM2, the putative dimeric partner of BES1 (25–27). BEH1 to 4 and BIM2 were up-regulated in the meristemoid-enriched mutant and BIM2 exhibited stomatal lineage-specific expression (Fig. 4, B and D). Epidermal expression of bes1-D (26) correlates with an increase in stomatal density, whereas a bes1 RNAi knockdown line (27) exhibited a trend toward lower stomatal density (Fig. 4E). This role in promoting stomatal development may be explained through the known repression of the BR biosynthetic genes by the BZR1 family (28). Thus, SPCH-mediated induction of BIN2 and repression of CPD (either directly or indirectly through the BZR1 family), leads to higher BIN2 activity and de-repression of SPCH, promoting accumulation of SPCH in active meristemoids (Fig. 4F). Overall, this feedback mechanism by SPCH would serve to reinforce differences between SPCH-expressing meristemoids and non-expressing neighbors which may be important for local patterning and coordinating the lineage with overall BR-mediated growth controls.
Fig. 4. Feedback regulation of brassinosteroid biosynthesis and signaling by SPCH.
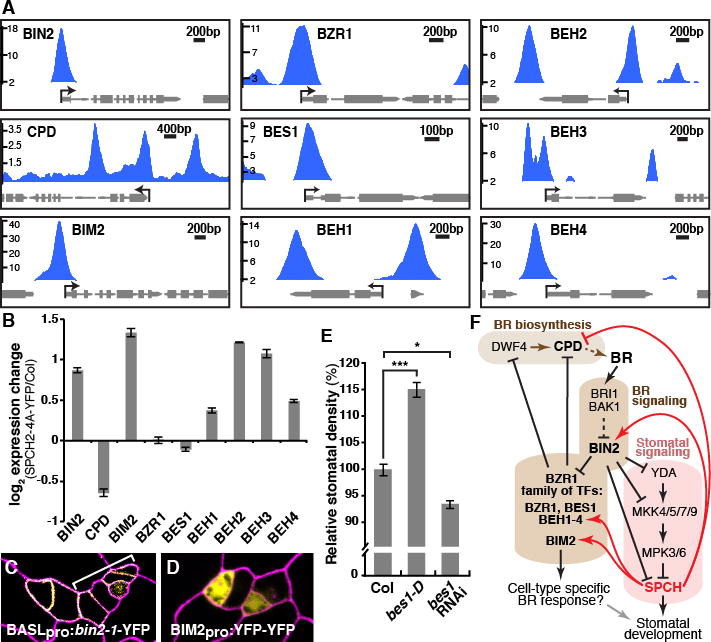
(A) ChIP-Seq profiles of select brassinosteroid (BR) pathway genes (labeled as in Fig. 2F). (B) RT-qPCR analysis of BR genes in 4-dpg SPCHpro:SPCH2-4A-YFP and Col seedlings. Values are means+/−SEM. (C & D) Confocal images of 3-dpg adaxial cotyledons with propidium iodide-stained cell outlines (purple). Stomatal lineage-specific expression of hyperactive BIN2 (yellow) induces lineage proliferation (bracket) (C). Stomatal lineage expression pattern of BIM2pro:YFP-YFP (yellow) (D). (E) Alteration of stomatal density in gain-of-function BES1pro:bes1-D and bes1-RNAi knockdown. *: p<0.05, ***: p<0.001 (Wilcoxon ranksum test). (F) Model of SPCH-BR pathway interactions. SPCH, a target of BR signaling, feeds back (positively, red arrows or negatively, red T-bars) upon transcription of multiple pathway members.
Here we revealed the broad influence of SPCH in stomatal lineage specification through MOBE-ChIP. This technique, which is based on simple scale increase, could be widely applicable in other tissues or organisms to obtain high-quality binding information about cell-type-specific regulators. The large number of SPCH-binding regions reported here is reminiscent of the behavior of the bHLH transcription factor MyoD, a master regulator of mammalian myogenesis, which associates with more than 30,000 regions in the human genome and is responsible for resetting global transcriptional and epigenetic states during development (29). Additional experiments are needed to establish definitively how often and by what mechanisms SPCH binding alters gene expression. However, our data that hundreds of genes, including those mediating abiotic and hormone responses, are directly regulated by SPCH supports previous functional studies (20, 22) that place SPCH in a critical position to integrate physiological and environmental information into a developmental program that optimizes leaf properties (stomatal density and size) for prevailing environments.
Supplementary Material
Acknowledgments
We thank Z.-Y. Wang (Carnegie) for the anti-YFP antibody and the bin2-1 allele; Y. Yin (ISU) for the BES1pro:bes1-D-GFP construct; J. Chory (SALK) for the bes1 RNAi line; and members of our laboratory for critical comments. Funding for this work was provided by NIH 1R01GM086632. OSL was a Croucher Fellow, KAD was supported by Cellular and Molecular Biology Training Program NIH5T32GM007276 and by an NSF graduate research fellowship, and JA was supported by the DAAD. DCB is a Gordon and Betty Moore Foundation Investigator of the Howard Hughes Medical Institute. The ChIP-Seq and RNA-Seq data in this study can be found in NCBI’s GEO repository (http://www.ncbi.nlm.nih.gov/geo/) as GSE57954. Supplement contains additional data.
Footnotes
References and Notes
- 1.Heidstra R, Sabatini S. Plant and animal stem cells: similar yet different. Nat Rev Mol Cell Biol. 2014;15:301–312. doi: 10.1038/nrm3790. [DOI] [PubMed] [Google Scholar]
- 2.Fong AP, Tapscott SJ. Skeletal muscle programming and re-programming. Curr Opin Genet Dev. 2013;23:568–573. doi: 10.1016/j.gde.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.García-Bellido A, de Celis JF. The complex tale of the achaete-scute complex: a paradigmatic case in the analysis of gene organization and function during development. Genetics. 2009;182:631–639. doi: 10.1534/genetics.109.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau OS, Bergmann DC. Stomatal development: a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development. 2012;139:3683–3692. doi: 10.1242/dev.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillitteri LJ, Dong J. Stomatal development in Arabidopsis. Arabidopsis Book. 2013;11:e0162. doi: 10.1199/tab.0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson S, et al. Generation of spatial patterns through cell polarity switching. Science. 2011;333:1436–1440. doi: 10.1126/science.1202185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacAlister CA, Ohashi-Ito K, Bergmann DC. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature. 2007;445:537–540. doi: 10.1038/nature05491. [DOI] [PubMed] [Google Scholar]
- 8.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007;445:501–505. doi: 10.1038/nature05467. [DOI] [PubMed] [Google Scholar]
- 9.Muiño JM, Kaufmann K, van Ham RC, Angenent GC, Krajewski P. ChIP-seq Analysis in R (CSAR): An R package for the statistical detection of protein-bound genomic regions. Plant Methods. 2011;7:11. doi: 10.1186/1746-4811-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiessl K, Muiño JM, Sablowski R. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc Natl Acad Sci USA. 2014;111:2830–2835. doi: 10.1073/pnas.1320457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaufmann K, et al. Orchestration of Floral Initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- 12.Dong J, MacAlister CA, Bergmann DC. BASL controls asymmetric cell division in Arabidopsis. Cell. 2009;137:1320–1330. doi: 10.1016/j.cell.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillitteri LJ, Peterson KM, Horst RJ, Torii KU. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell. 2011;23:3260–3275. doi: 10.1105/tpc.111.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanaoka MM, et al. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–1700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 16.Shpak ED, McAbee JM, Pillitteri LJ, Torii KU. Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science. 2005;309:290–293. doi: 10.1126/science.1109710. [DOI] [PubMed] [Google Scholar]
- 17.Hunt L, Gray JE. The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol. 2009;19:864–869. doi: 10.1016/j.cub.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 18.Malcos JL, Cyr RJ. An ungrouped plant kinesin accumulates at the preprophase band in a cell cycle-dependent manner. Cytoskeleton (Hoboken) 2011;68:247–258. doi: 10.1002/cm.20508. [DOI] [PubMed] [Google Scholar]
- 19.Rasmussen CG, Humphries JA, Smith LG. Determination of symmetric and asymmetric division planes in plant cells. Annu Rev Plant Biol. 2011;62:387–409. doi: 10.1146/annurev-arplant-042110-103802. [DOI] [PubMed] [Google Scholar]
- 20.Le J, et al. Auxin transport and activity regulate stomatal patterning and development. Nat Comms. 2014;5:3090. doi: 10.1038/ncomms4090. [DOI] [PubMed] [Google Scholar]
- 21.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–422. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gudesblat GE, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14:548–554. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka Y, Nose T, Jikumaru Y, Kamiya Y. ABA inhibits entry into stomatal-lineage development in Arabidopsis leaves. Plant J. 2013;74:448–457. doi: 10.1111/tpj.12136. [DOI] [PubMed] [Google Scholar]
- 24.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 27.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 28.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lampard GR, MacAlister CA, Bergmann DC. Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science. 2008;322:1113–1116. doi: 10.1126/science.1162263. [DOI] [PubMed] [Google Scholar]
- 31.Roeder AHK, et al. Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 2010;8:e1000367. doi: 10.1371/journal.pbio.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa T, et al. Development of R4 gateway binary vectors (R4pGWB) enabling high-throughput promoter swapping for plant research. Biosci Biotechnol Biochem. 2008;72:624–629. doi: 10.1271/bbb.70678. [DOI] [PubMed] [Google Scholar]
- 33.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borner GHH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubo M, et al. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 37.Clough SJ. Floral dip: agrobacterium-mediated germ line transformation. Methods Mol Biol. 2005;286:91–102. doi: 10.1385/1-59259-827-7:091. [DOI] [PubMed] [Google Scholar]
- 38.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 39.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nature Protocols. 2008;3:1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- 40.Li R, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 41.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cairns J, et al. BayesPeak–an R package for analysing ChIP-seq data. Bioinformatics. 2011;27:713–714. doi: 10.1093/bioinformatics/btq685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic acids research. 2010;38:W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 46.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B. QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics. 2008;9:465. doi: 10.1186/1471-2105-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 50.Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development. 2010;137:447–455. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- 52.Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–1497. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 53.Hara K, et al. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009;50:1019–1031. doi: 10.1093/pcp/pcp068. [DOI] [PubMed] [Google Scholar]
- 54.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Maoileidigh DS, et al. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell. 2013;25:2482–2503. doi: 10.1105/tpc.113.113209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chinnusamy V, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakai T, et al. Armadillo repeat-containing kinesins and a NIMA-related kinase are required for epidermal-cell morphogenesis in Arabidopsis. Plant J. 2008;53:157–171. doi: 10.1111/j.1365-313X.2007.03327.x. [DOI] [PubMed] [Google Scholar]
- 58.Khan M, et al. Brassinosteroid-regulated GSK3/Shaggy-like Kinases Phosphorylate Mitogen-activated Protein (MAP) Kinase Kinases, Which Control Stomata Development in Arabidopsis thaliana. J Biol Chem. 2013;288:7519–7527. doi: 10.1074/jbc.M112.384453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. The Plant Journal. 2011;65:634–646. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


