Abstract
Hepatitis E virus (HEV) is a serious public health problem. The commonly used tests that are specific for current HEV infection diagnosis include the detection of anti-HEV IgM and HEV RNA. Here, we report an improved enzyme-linked immunosorbent assay (ELISA) method for HEV antigen detection with a linear range equivalent to 6.3 × 103 to 9.2 × 105 RNA copies per ml. The monoclonal antibody (MAb) 12F12, a high-ability MAb that binds HEV virus, was selected as the capture antibody from a panel of 95 MAbs. The positive period of HEV antigenemia in infected monkeys using this test was, on average, 3 weeks longer than previously reported and covered the majority of the acute phase. The positive detection rates of IgM, RNA, and new antigen from the first serum samples collected from 16 confirmed acute hepatitis E patients were 81% (13/16), 81% (13/16), and 100% (16/16), respectively. In three patients, the initial serum specimens that tested negative for IgM, despite the presence of symptoms of acute hepatitis and elevated alanine aminotransferase (ALT) levels, were positive for HEV antigen and HEV RNA. In contrast, the serum samples of the three RNA-negative patients were antigen positive (and IgM positive), possibly due to the degradation of HEV nucleic acids. Our results suggest that this new antigen detection method has acceptable concordance with RNA detection and could serve as an important tool for diagnosing acute hepatitis E.
INTRODUCTION
Hepatitis E is an enterically transmitted viral hepatitis caused by hepatitis E virus (HEV) infection (1). Hepatitis E is a serious public health problem in many countries (especially developing countries), with a mortality rate of approximately 20 to 25% among pregnant women (2). HEV is a 34-nm, nonenveloped, and icosahedral virus (3) with a 7.2-kb positive-sense single-stranded RNA genome containing three open reading frames. Open reading frame 2 (ORF2) (660 amino acids) encodes the major viral capsid (4). Mammalian HEV is divided into four genotypes with distinct geographic distributions and prevalences (5, 6).
Most patients with acute hepatitis E infection present with typical acute hepatitis symptoms, such as jaundice and dark urine. Typical biochemical changes in acute HEV patients include increased serum levels of alanine aminotransferase and aspartate aminotransferase (ALT and AST, respectively) and bilirubin; however, these factors are not specific for hepatitis E, as increases also occur due to other viral and nonviral forms of liver injury. The most commonly used tests specific for diagnosing HEV infection detect anti-HEV IgM and HEV RNA. In acute hepatitis E patients, anti-HEV IgM can typically be detected within 3 to 4 days after the onset of jaundice and can persist for an average of 5 months (7). The presence of anti-HEV IgM provides evidence of recent HEV infection; however, its short detection period indicates that anti-HEV IgM is unsuitable as a single marker for current infection in acute hepatitis E patients. In contrast, the detection of HEV RNA provides a specific and highly sensitive approach to the diagnosis of current HEV infection. However, RNA detection is technically complex, cumbersome, costly, and prone to contamination. Moreover, as a type of naked RNA virus, the virions of HEV are unstable (8), and the nucleic acids are easily degraded.
The laboratory diagnosis of the pathogen relies on the direct detection of one of its components (i.e., nucleic acids or proteins). The detection of the HEV capsid protein has been attempted using a sandwich enzyme immunoassay (9). An analysis of serial samples from HEV-infected rhesus monkeys showed that the HEV capsid antigen could be detected almost simultaneously with the RNA in stool, but the capsid antigen disappeared 2 to 3 weeks earlier than the RNA. Furthermore, the period of antigen positivity was shorter in the serum than in the stool. Thus, the period of antigen positivity did not match the period of ALT elevation in most rhesus monkeys.
In this study, we improved HEV antigen detection by using a different antibody pair in a sandwich enzyme immunoassay. Our results show that the novel method has good concordance with RNA detection, and the positive period for the serum viral antigen was, on average, 3 weeks longer than was previously reported. By using this improved high-sensitivity HEV antigen detection method, we conducted this study to further define the role of HEV antigen detection in acute hepatitis E infection through a comparative analysis of current HEV infection-related markers, such as the HEV antigen, HEV RNA, anti-HEV IgM, and elevated ALT levels.
MATERIALS AND METHODS
Monoclonal antibodies and hepatitis E virus.
A panel of 95 monoclonal antibodies (MAbs) that recognize the HEV capsid were obtained using a standard murine MAb preparation protocol (10) from mice immunized with recombinantly expressed HEV capsid. The recombinant protein represents a portion of the HEV capsid protein encompassed within recombinant particle protein p239 (amino acids 368 to 606) (11) and recombinant dimer protein E2 (amino acids 394 to 606) (12). Monoclonal antibody (MAb) no. 4 was donated by Youchun Wang (9). The viruses were isolated from stool samples from rhesus monkeys infected with HEV genotype 1 virus (strain Xinjiang), genotype 3 virus (strain JRC-HE3), and genotype 4 virus (strain Ch-S-1).
Detection of HEV RNA by quantitative reverse transcriptase PCR.
HEV RNA was purified from 50 μl of each stool and serum sample from HEV-infected rhesus monkeys and clinical serum specimens from confirmed hepatitis E cases. The copy number of HEV RNA was determined using a quantitative real-time reverse transcriptase PCR (RT-PCR) assay, as previously reported (13). A CFX96 real-time system and C1000 thermal cycler device (Bio-Rad, Inc., Hercules, CA) were used for all real-time RT-PCR tests. For the generation of standard quantitation curves, the threshold cycle (CT) values were plotted as a function of the input HEV viral copy numbers. The copy numbers were determined by calibrating the concentration of the plasmid standard.
Antigen detection protocol for the sandwich ELISA with 12F12 as the capture Ab.
A sandwich enzyme-linked immunosorbent assay (ELISA) was established by coating microtiter plates with MAb 12F12 and detecting the captured antigen with horseradish peroxidase (HRP)-conjugated MAb no. 4. The microtiter plates were coated with 500 ng per well of MAb 12F12 and then washed once with phosphate-buffered saline (PBS) containing 0.5% (wt/vol) Tween 20 (PBST) and blocked with 200 μl of PBS containing 2% (wt/vol) bovine serum albumin (BSA). A total of 50 μl of serum or stool sample was added and incubated for 60 min at 37°C without washing. Next, the plates were incubated with 100 μl per well of MAb 4-HRP solution at 37°C for 30 min and then washed five times with PBST. Subsequently, 100 μl of tetramethylbenzidine substrate solution was added and incubated for 15 min at 37°C. The absorbance was measured at 450 nm, with 620 nm as a background, using a microplate reader (Autobio, Zhengzhou, China).
Analysis of the corresponding relationship between novel antigen detection and RNA detection.
Two stool samples from rhesus monkeys infected with each genotype and exhibiting high HEV viral loads were evaluated in parallel. After a preliminary evaluation and adjustment, 2-fold serial dilutions were prepared for all six specimens prior to simultaneous testing for RNA and antigen. The results were analyzed with the GraphPad Prism software. A curve was fitted for nonlinear regression with log[agonist] versus response − variable slope.
Analysis of samples from rhesus monkeys infected with HEV genotypes 1, 3, and 4.
Rhesus monkeys were infected with virus, and serial samples (including those from feces and serum) were collected as previously described (14). Briefly, the rhesus monkeys were inoculated with 1E07 copies of HEV. The serum samples were analyzed for anti-HEV antibodies (Wantai Biopharm, Beijing, China), HEV RNA, ALT (Wantai Biopharm), and HEV antigen. The results of the HEV antigen, anti-HEV IgM, and anti-HEV IgG assays are shown as a positive-to-negative ratio (P/N). The baseline serum samples from three monkeys served as negative controls. The mean value of the baseline serum samples from the three monkeys was <0.05; thus, 0.05 was used as the negative control. A P/N value of ≥2.1 was defined as positive.
Collection and analysis of a series of serum samples from acute hepatitis E patients.
Samples were collected from acute hepatitis E patients in Dongtai, Jiangsu Province (China) from June 2011 to May 2012. The probable cases of acute hepatitis were defined as patients who suffered from fatigue and/or loss of appetite for ≥3 days and who had an elevated ALT level of ≥2.5-fold above the upper limit of normal (ULN). Serum samples from the acute hepatitis patients were collected between 2 and 12 days after the onset of symptoms and at ≥1 follow-up time point. The serum samples were tested for ALT, anti-HEV antibodies, and HEV RNA (using the methods described above). The anti-HEV IgG level of each sample was determined using a WHO reference serum (15). The confirmed cases of acute hepatitis E were defined as those meeting at least two of the following criteria: (i) the presentation of HEV RNA within 35 days after onset, (ii) IgM seroconversion within 70 days after onset, or (iii) a ≥4-fold increase in IgG levels in paired sera. Among 41 possible hepatitis E patients, 32 patients were subjected to follow-up, and a total of 16 cases were confirmed acute hepatitis E cases. The confirmed cases were included in this study. The serum specimens from the confirmed acute HEV patients were further analyzed for HEV antigen using the improved sandwich ELISA. The cutoff value of the antigen sandwich ELISA was determined using 1,424 non-HEV (normal ALT levels, HEV RNA negative, and anti-HEV antibody negative) volunteer blood donor serum samples from the Xiamen blood station. Twenty-six RNA-positive serum specimens from acute hepatitis E patients were also collected to analyze the specificity of the sandwich ELISA. Sixteen serum samples from hepatitis A patients, 65 serum samples from hepatitis B patients, and 46 serum samples from hepatitis C patients were also included to analyze the specificity of the sandwich ELISA. Hepatitis A patients were defined as hepatitis patients who were hepatitis A virus (HAV) IgM positive and negative for hepatitis B virus (HBV), hepatitis C virus (HCV), and HEV markers. Hepatitis B patients were defined as hepatitis patients who were HBsAg positive and negative for HAV, HCV, and HEV markers. Hepatitis C patients were defined as hepatitis patients who were HCV IgG positive and negative for other viral hepatitis markers.
RESULTS
Calibration curve of the sandwich ELISA.
A sandwich ELISA was established by coating microtiter plates with MAb 12F12 and using HRP- conjugated MAb no. 4 for detection. The paired MAbs were screened from a panel containing 96 anti-HEV MAbs. The selection process for this pair of MAbs in establishing the sandwich ELISA is shown in Fig. S1 in the supplemental material.
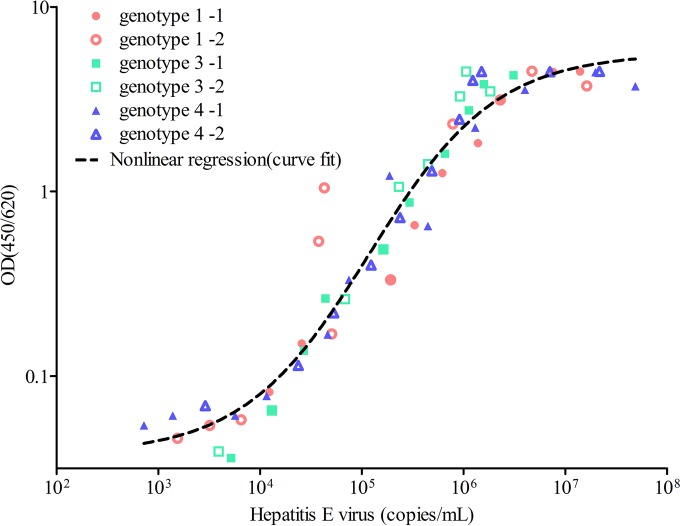
Most human hepatitis E virus strains belong to genotypes 1, 3, and 4; human genotype 2 has been detected less frequently and is responsible for a single epidemic in Mexico. Thus, we tested the improved ELISA method on viruses from genotypes 1, 3, and 4. To investigate the relationship between antigen detection and RNA viral load, we conducted simultaneous tests on serially diluted specimens. The calibration curves of the concentration of antigen versus viral load for the sandwich ELISA were established using the serially diluted genotype 1, 3, and 4 virions. The results showed a direct correlation between the antigen test and RNA viral load in the range of 6.3 × 103 to 9.2 × 105 copies/ml of viral particles (Fig. 1). The strong correlation between the optical density of the antigen and RNA viral load for all three genotypes of hepatitis E supported the finding that the primary MAb recognized an epitope common to all three HEV genotypes.
FIG 1.
The standard curves of the sandwich ELISA were established using serially diluted stool samples from HEV-infected rhesus macaques (genotype 1, pink [sample 1, closed circle; sample 2, open circle]; genotype 3, blue [sample 1, closed square; sample 2, open square]; genotype 4, purple [sample 1, closed triangle; sample 2, open triangle]) as antigen targets. Serially diluted HEV viral loads were quantified by real-time RT-PCR. The results were analyzed with the GraphPad Prism software. A curve was fitted for nonlinear regression. The sandwich ELISA had similar detection abilities for viruses with different genotypes. The range of correlation between the viruses was 6.3 × 103 to 9.2 × 105 copies/ml.
Diagnostic performance of the novel antigen detection method on serial samples from rhesus monkeys inoculated with HEV.
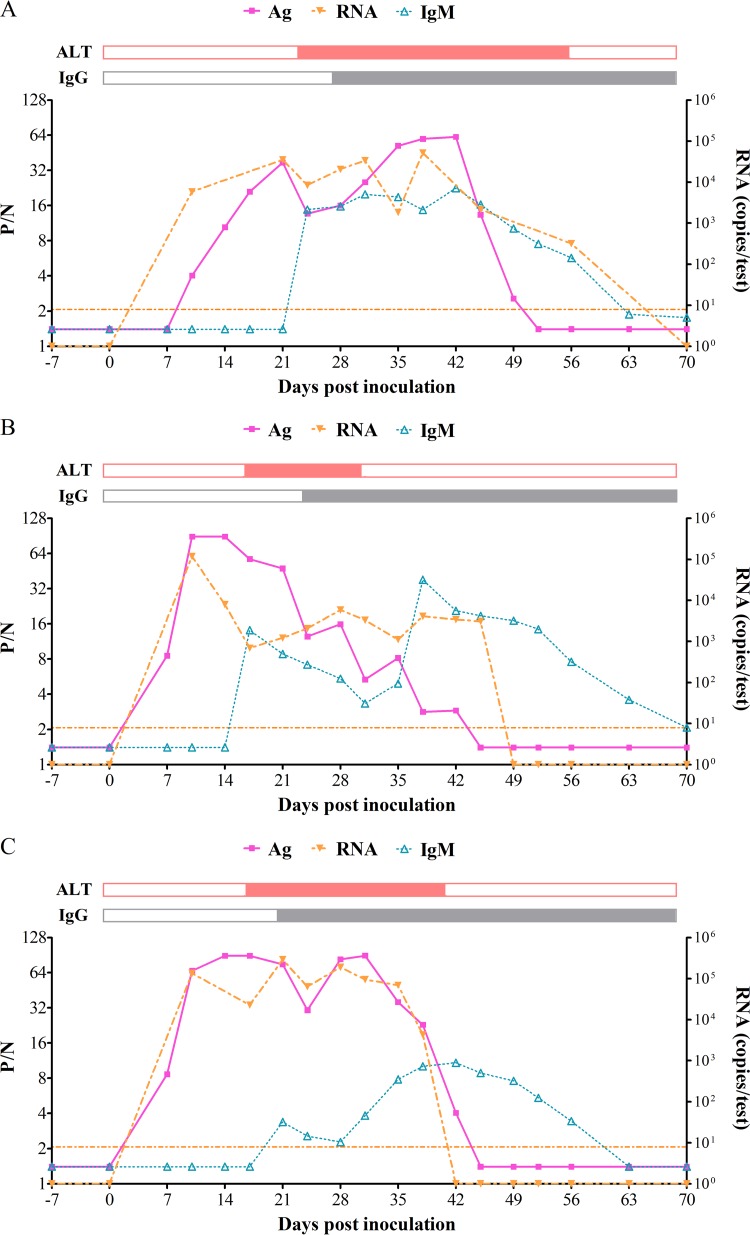
To identify the period of HEV antigen detection in the sera, the series serum samples from three monkeys inoculated with HEV genotypes 1, 3, and 4 were tested for HEV antigen using a novel detection method; simultaneously, we determined the levels of HEV RNA, anti-HEV antibodies, and ALT in these samples.
As shown in Fig. 2, the HEV antigen had good concordance with RNA viral load. Both antigen and nucleic acids were detectable at 7 to 10 days postinoculation (p.i.) and remained positive for 5 to 7 weeks in the serum samples from all three monkeys. These results are in contrast with those of an earlier study in which antigen positivity was observed for 2 to 4 weeks in serum, while RNA in the stool was detectable for 5 to 8 weeks. Anti-HEV IgM seroconversion and ALT elevation were detected 1 to 2 weeks later, with the samples becoming positive for anti-HEV IgG at 21 to 28 days p.i. Both antigen and RNA became undetectable approximately 3 weeks after IgG-positive seroconversion.
FIG 2.
The relationship between HEV antigen, antibodies, RNA, and ALT on serial serum samples collected from rhesus monkeys infected with genotype 1 (A), genotype 3 (B), and genotype 4 (C) HEV. The results of anti-HEV antibody and antigen detection were recorded as positive-to-negative ratio (P/N). A P/N value of ≥2.1 was defined as positive. An abnormal ALT level was defined as ≥40 U/liter. The results of ALT and anti-HEV IgG are shown as closed and open bars (above each panel), respectively. The closed bars indicate anti-HEV IgG positivity/abnormal ALT levels, and the open bars indicate anti-HEV IgG negativity/normal ALT levels.
The HEV antigen and RNA viral load were also measured in fecal samples; the fecal HEV antigen and RNA levels were similar to those detected in the serum (see Fig. S2 in the supplemental material). HEV antigen and HEV RNA became positive and then undetectable almost simultaneously. HEV antigen positivity and RNA positivity were observed from 7 to 56 days p.i.
Determination of the cutoff value and specificity of the sandwich ELISA.
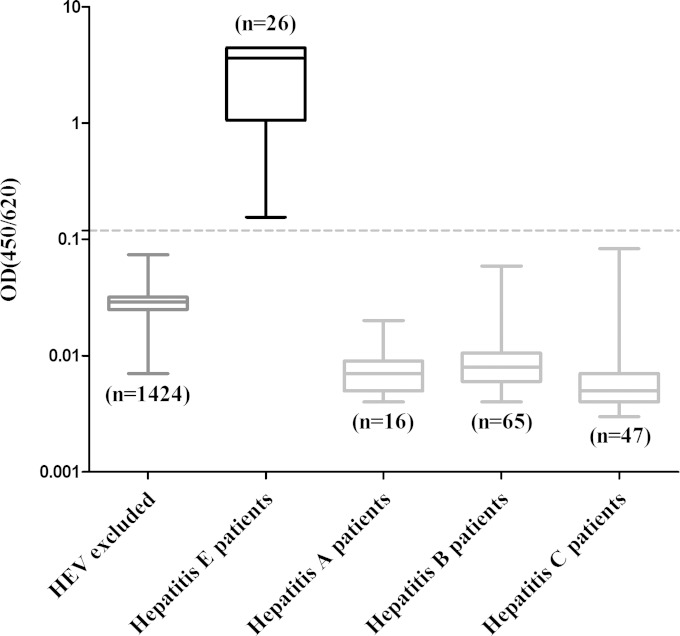
The cutoff value was determined before the sandwich ELISA was performed in clinical serum samples. Using 1,424 non-HEV (normal ALT levels, HEV RNA negative, and anti-HEV antibody negative) volunteer blood donor serum samples from the Xiamen blood station (see Fig. S3 in the supplemental material), the cutoff value of the antigen sandwich ELISA was determined to be a mean optical density plus 8 standard deviations (SDs) of 0.12. The calculated specificity was 100%, and when the cutoff value was set at 0.12, none of the serum samples from the healthy population were HEV antigen positive (Fig. 3). Twenty-six RNA-positive serum specimens from acute hepatitis E patients were positive for HEV antigen. Sixteen serum samples from hepatitis A patients, 65 serum samples from hepatitis B patients, and 46 serum samples from hepatitis C patients were collected and used as disease controls; all hepatitis A, B, and C serum samples were negative for HEV antigen detection (Fig. 3).
FIG 3.
The cutoff value and the specificity of the antigen sandwich ELISA were determined using 1,424 non-HEV specimens and 26 RNA-positive serum specimens collected from acute hepatitis E patients. The 1,424 non-HEV serum specimens were defined as those with normal ALT levels and were negative for HEV RNA and anti-HEV antibodies. The cutoff value was set as the mean optical density plus 8 SDs (0.12). The dotted line represents the cutoff value of the antigen. Sixteen serum specimens from hepatitis A patients, 65 serum specimens from hepatitis B patients, and 46 serum specimens from hepatitis C patients were included in this analysis to evaluate the specificity of the method. The detection results for each group are shown as the range (whiskers), interquartile range (boxes), and median (line within boxes).
Relationship between HEV antigen, HEV RNA, anti-HEV IgM, and ALT in serial clinical serum specimens collected from confirmed hepatitis E cases.
To compare the capsid antigen with other acute hepatitis E-related markers, we used a series of serum specimens from 16 acute hepatitis E patients for further analysis. These 16 acute hepatitis E patients were identified during June 2011 to May 2012 in Dongtai, Jiangsu Province, China. Potential acute hepatitis patients meeting at least two of three acute hepatitis E-related criteria (i.e., HEV RNA positivity, anti-HEV IgM positivity, and a 4-fold increase in anti-HEV IgG) were considered to be confirmed acute hepatitis E patients. The presence of HEV antigen in 47 serum specimens from these 16 acute hepatitis E patients was tested for and then compared with HEV RNA, anti-HEV antibody, and ALT data.
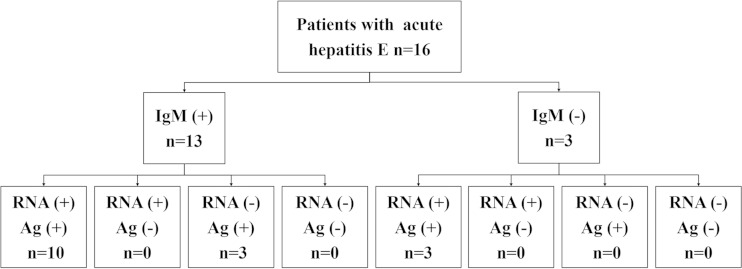
As shown in Fig. 4, the first serum samples collected from all 16 patients were identified as HEV antigen positive by the novel detection method, while only 13 of these samples were anti-HEV IgM positive. In the three IgM-negative serum samples, HEV RNA and HEV antigen were positively identified, indicating the presence of the pathogen in the first serum samples collected from these three acute hepatitis E patients, despite an undetectable antibody response. The follow-up serum samples from these three patients showed both an anti-HEV IgM seroconversion and a 4-fold elevation in anti-HEV IgG, which confirmed that these three patients were infected with HEV. To our surprise, three of the 16 serum samples collected in the initial round were HEV RNA negative but HEV antigen and anti-HEV IgM positive. These three serum samples also tested RNA negative when using a reverse transcriptase PCR (RT-PCR) assay with different primers (16).
FIG 4.
A schematic of the diagnostic tests conducted on the first serum samples collected from confirmed acute hepatitis E cases, with the results obtained for anti-HEV IgM, HEV RNA, and HEV antigen (Ag) testing. Sixteen patients were included. The first serum samples collected from 13 patients were positive for RNA detection. The first sera collected from three patients were negative for anti-HEV IgM detection. All samples were HEV antigen positive.
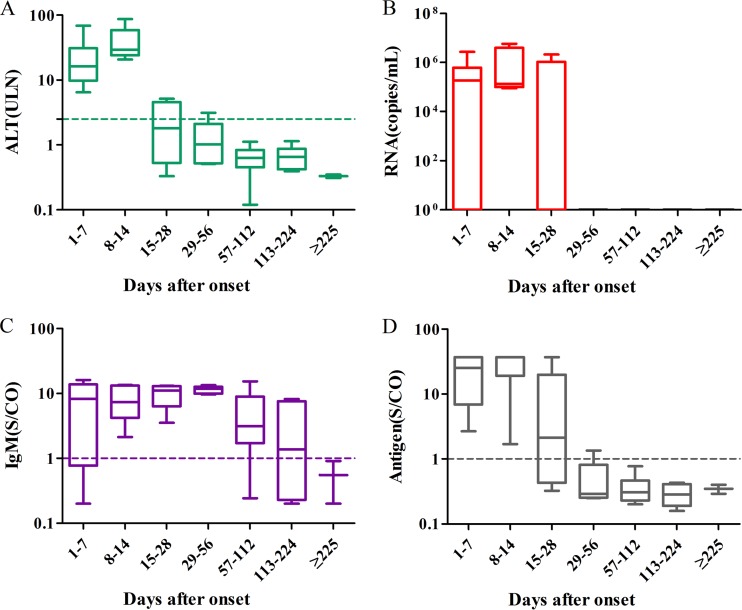
Figure 5 presents the ALT, anti-HEV IgM, and pathogen dynamics observed during the progression of the illness. The normalization of ALT levels and virus clearance were achieved in most patients within 28 days after the onset of symptoms. All HEV RNA-positive serum specimens but one were collected during the 14 days after the onset of symptoms.
FIG 5.
The dynamics of ALT, RNA, IgM, and antigen in acute hepatitis during the progression of illness. Serum samples were collected from 16 confirmed acute hepatitis E cases within the first week of the onset of symptoms (n = 11) and in subsequent intervals after the onset of symptoms, from 8 to 14 days (n = 5), from 15 to 28 days (n = 5), from 29 to 56 days (n = 5), from 57 to 112 days (n = 13), from 113 to 224 days (n = 6), and after >225 days (n = 2). The dotted lines represent the cutoff levels of ALT, anti-HEV IgM, and HEV antigen. The levels of ALT (A), viral RNA (B), anti-HEV IgM (C), and antigen (D) of the different groups of serum samples are shown as the range (whiskers), interquartile range (boxes), and median (line within the boxes) values.
HEV antigen positivity was observed mainly during the 28 days after the onset of symptoms. Periods of HEV antigen positivity and HEV RNA positivity were present in the early phase and were similar to the period of ALT elevation. This finding confirms that the pathogen indicators are related to the acute phase of hepatitis E infection and are accompanied by elevated ALT levels and clinical symptoms. Most patients were positive for anti-HEV IgM at the time of first serum collection, with anti-HEV IgM levels reaching a peak at 29 to 56 days after the onset of symptoms. HEV IgM was negative in all samples collected 225 days after the onset of symptoms.
DISCUSSION
HEV is one of the major causes of acute hepatitis worldwide (17). Several diagnostic assays for anti-HEV antibodies are available, but they often provide discordant results. The presence of anti-HEV IgG represents either recent or remote exposure and is a marker of anamnesis exposure. The detection of anti-HEV-IgM is currently regarded as the routine method for diagnosing acute HEV infection, despite the fact that IgM can persist for an average of 5 months in HEV-infected patients, while the acute phase of hepatitis E lasts for several weeks (18). Therefore, anti-HEV IgM indicates recent but not current infection. To the best of our knowledge, HEV RNA is considered the gold standard for the detection of both acute and chronic current infection. However, RNA detection is difficult to control and is available only in specialized laboratories. HEV antigen, another indicator of current pathogen presence, is better correlated with current HEV infection (19) and markers of liver function (20) than with anti-HEV IgM. Hence, a good antigen reagent will play a key role in the diagnosis of acute hepatitis E infection.
In this study, we established an improved sandwich ELISA for detecting HEV antigen. In an analysis of serial stool and serum samples from HEV-infected rhesus monkeys using this new method, we found that the presence of HEV antigen was in concordance with the presence of RNA. In all three monkeys, both HEV antigen and RNA were detectable in the serum samples for 5 to 7 weeks, which represents an average of 3 weeks longer than what was previously reported (9). The detectable periods for HEV antigen and RNA in the serum samples were similar to those in feces, which also differed from the results based on the previously reported method (9). In summary, our novel method showed good sensitivity for detection in serum samples that may be attributed to the high affinity of MAbs for the HEV antigen used in the new method. The MAbs have the ability to compete for the viral antigen with host-specific antibodies, which form a complex with the antigen and affect antigen detection in serum samples. With this improvement, viral antigens were still detectable, even when an anti-HEV antibody response was present. Moreover, the antigen-positive phases in the serum samples almost completely overlapped with the ALT elevation periods. This differed from previously reported results in which the antigen-positive periods in sera showed poor agreement with elevated ALT levels in most HEV-infected rhesus monkeys. Because ALT elevation is a representative indicator for acute hepatitis and is highly correlated with related symptoms, the novel antigen detection method truly indicates current infection with HEV and the acute phase of hepatitis E.
The novel ELISA method showed positive results using the first serum sample collected from 16 confirmed acute hepatitis E patients. This indicates that the HEV antigen is a valuable indicator of acute hepatitis E and that this novel antigen detection method has a high sensitivity for diagnosing hepatitis E virus infection. Notably, three of these 16 first serum samples were HEV RNA negative but HEV antigen and anti-HEV IgM positive. Moreover, a 4-fold increase in anti-HEV IgG was observed in serial serum samples collected from these three patients. This phenomenon may be attributable to the degradation of HEV RNA during the storage and transportation steps, which is an additional limitation of the extensive use of RNA for detection.
From the results of the samples from the acute hepatitis E patients, we also found that the first serum samples collected from three patients were anti-HEV IgM negative but pathogen positive (both RNA and antigen positive). This finding indicated that these three patients were in the early stages of infection and that antibody responses to the current infection were undetectable. The typical HEV infection model shows that acute hepatitis E is accompanied by an increase in anti-HEV IgM concentrations (18). However, in these three patients, ALT elevation and acute hepatitis symptoms were present without significant IgM seroconversion, suggesting that HEV infection also resulted in liver injury and other related diseases before the appearance of anti-HEV IgM. Due to the lack of genotype 2 clinical samples and isolated viruses, this study showed that this improved method has good sensitivity for viral and clinical serum samples with genotypes 1, 3, and 4. However, the use of genotype 2 recombinant capsid E2 as an alternative for detection suggested that this sandwich ELISA possesses the same detection ability for recombinant capsids from all 4 genotypes (see Fig. S4 in the supplemental material).
In conclusion, this study developed a novel antigen detection method with good sensitivity for both pathogen detection and indication of the acute phase of hepatitis E. An analysis based on this novel method further confirmed that the HEV antigen, as an indicator of the pathogen, is highly consistent with the presence of viral RNA and coincides with acute hepatitis E infection. Considering the instability of HEV RNA, this novel antigen detection method should be valuable for widespread use as a hepatitis E diagnostic tool. HEV antigen detection based on this novel method will play an important role in the diagnosis of acute hepatitis E and screening for HEV-positive blood donors, both in the acute phase and during the period after seroconversion to anti-HEV.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Chinese National High-Tech R&D Program (863 program; grant 2011AA02A101), the Xiamen science and technology platform project (3502Z20131001), and the Xiamen science and technology plan project (3502Z201410045).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01853-14.
REFERENCES
- 1.Reyes GR, Purdy MA, Kim JP, Luk K-C, Young LM, Fry KE, Bradley DW. 1990. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science 247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 2.Jaiswal S, Jain A, Naik G, Soni N, Chitnis D. 2001. Viral hepatitis during pregnancy. Int J Gynaecol Obstet 72:103–108. doi: 10.1016/S0020-7292(00)00264-2. [DOI] [PubMed] [Google Scholar]
- 3.Emerson SU, Purcell RH. 2003. Hepatitis E virus. Rev Med Virol 13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 4.Tam AW, Smith MM, Guerra ME, Huang C-C, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell R, Emerson S. 2008. Hepatitis E: an emerging awareness of an old disease. J Hepatol 48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Kamar N, Bendall R, Legrand-Abravanel F, Xia NS, Ijaz S, Izopet J, Dalton HR. 2012. Hepatitis E. Lancet 379:2477–2488. doi: 10.1016/S0140-6736(11)61849-7. [DOI] [PubMed] [Google Scholar]
- 7.Myint KSA, Endy TP, Shrestha MP, Shrestha SK, Vaughn DW, Innis BL, Gibbons RV, Kuschner RA, Seriwatana J, Scott RM. 2006. Hepatitis E antibody kinetics in Nepalese patients. Trans R Soc Trop Med Hyg 100:938–941. doi: 10.1016/j.trstmh.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Zafrullah M, Khursheed Z, Yadav S, Sahgal D, Jameel S, Ahmad F. 2004. Acidic pH enhances structure and structural stability of the capsid protein of hepatitis E virus. Biochem Biophys Res Commun 313:67–73. doi: 10.1016/j.bbrc.2003.11.088. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Li X, Li Z, Harrison TJ, Chong H, Qiao S, Huang W, Zhang H, Zhuang H, Wang Y. 2006. Detection of HEV antigen as a novel marker for the diagnosis of hepatitis E. J Med Virol 78:1441–1448. doi: 10.1002/jmv.20717. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Gu Y, Ge SX, Li SW, He ZQ, Huang GY, Zhuang H, Ng MH, Xia NS. 2005. Analysis of hepatitis E virus neutralization sites using monoclonal antibodies directed against a virus capsid protein. Vaccine 23:2881–2892. doi: 10.1016/j.vaccine.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 11.Li SW, Zhang J, Li YM, Ou SH, Huang GY, He ZQ, Ge SX, Xian YL, Pang SQ, Ng MH, Xia NS. 2005. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine 23:2893–2901. doi: 10.1016/j.vaccine.2004.11.064. [DOI] [PubMed] [Google Scholar]
- 12.Zhang J-Z, Ng MH, Xia NS, Lau SH, Che XY, Chau TN, Lai ST, Im SWK. 2005. Conformational antigenic determinants generated by interactions between a bacterially expressed recombinant peptide of the hepatitis E virus structural protein. J Med Virol 64:125–132. doi: 10.1002/jmv.1027. [DOI] [PubMed] [Google Scholar]
- 13.Jothikumar N, Cromeans TL, Robertson BH, Meng X, Hill VR. 2006. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131:65–71. doi: 10.1016/j.jviromet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Ge SX, Huang GY, Li SW, He ZQ, Wang YB, Zheng YJ, Gu Y, Ng MH, Xia NS. 2003. Evaluation of antibody-based and nucleic acid-based assays for diagnosis of hepatitis E virus infection in a rhesus monkey model. J Med Virol 71:518–526. doi: 10.1002/jmv.10523. [DOI] [PubMed] [Google Scholar]
- 15.Zhu F-C, Zhang J, Zhang X-F, Zhou C, Wang Z-Z, Huang S-J, Wang H, Yang C-L, Jiang H-M, Cai J-P. 2010. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet 376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]
- 16.Li R-C, Ge S-X, Li Y-P, Zheng Y-J, Nong Y, Guo Q-S, Zhang J, Ng M-H, Xia N-S. 2006. Seroprevalence of hepatitis E virus infection, rural southern People's Republic of China. Emerg Infect Dis 12:1682–1688. doi: 10.3201/eid1211.060332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalton HR, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis 8:698–709. doi: 10.1016/S1473-3099(08)70255-X. [DOI] [PubMed] [Google Scholar]
- 18.Hoofnagle JH, Nelson KE, Purcell RH. 2012. Hepatitis E. N Engl J Med 367:1237–1244. doi: 10.1056/NEJMra1204512. [DOI] [PubMed] [Google Scholar]
- 19.Gupta E, Pandey P, Pandey S, Sharma MK, Sarin SK. 2013. Role of hepatitis E virus antigen in confirming active viral replication in patients with acute viral hepatitis E infection. J Clin Virol 58:374–377. doi: 10.1016/j.jcv.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhao C, Li L, Harrison TJ, Wang Q, Song A, Fan J, Ma H, Zhang C, Wang Y. 2009. Relationships among viral diagnostic markers and markers of liver function in acute hepatitis E. J Gastroenterol 44:139–145. doi: 10.1007/s00535-008-2281-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.