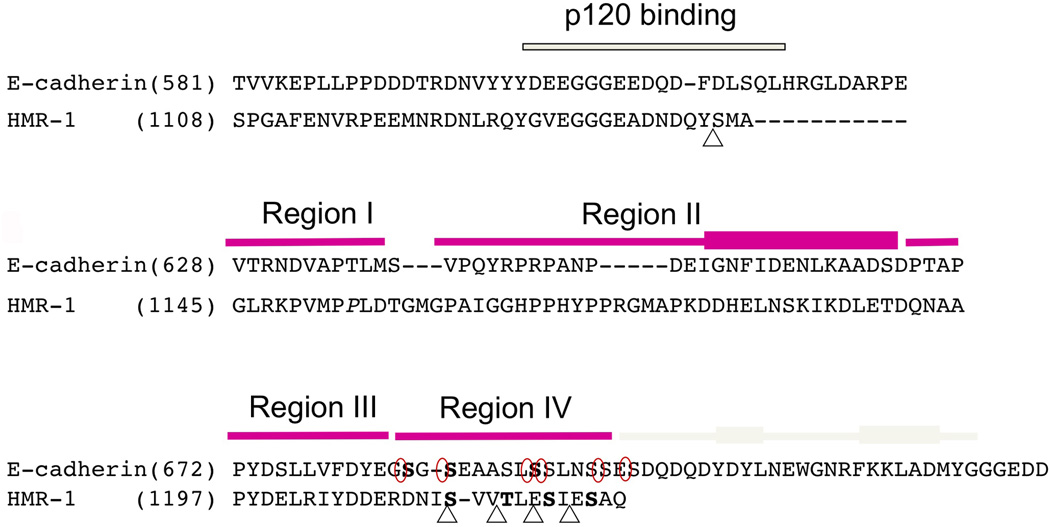
Figure 2. Sequence alignment of the cytoplasmic tails of mouse E-cadherin and HMR-1 reveals key conserved residues.
Juxtamembrane region including p120 (JAC-1 in C. elegans) binding site was aligned base on the sequence homology and β-catenin (HMP-2) binding region was aligned based on the crystal structures of pEcyto and pHMR-1cyto80. HMP-2 binding domain is divided into four regions and each region is indicated with magenta bar on top of the sequences. The C-terminal cap region is only present in Ecyto and is shown with grey colored bar. The thickened rectangles represent α-helices. Five CKI-phosphorylated sites in HMR-1cyto are marked with triangle and six phosphorylation sites by GSK-3β and CKII in Ecyto are circled. Phosphorylated residues observed in the structures are written in bold (3 residues in pEcyto and 4 in pHMRcyto80). The starting residue numbers are indicated.