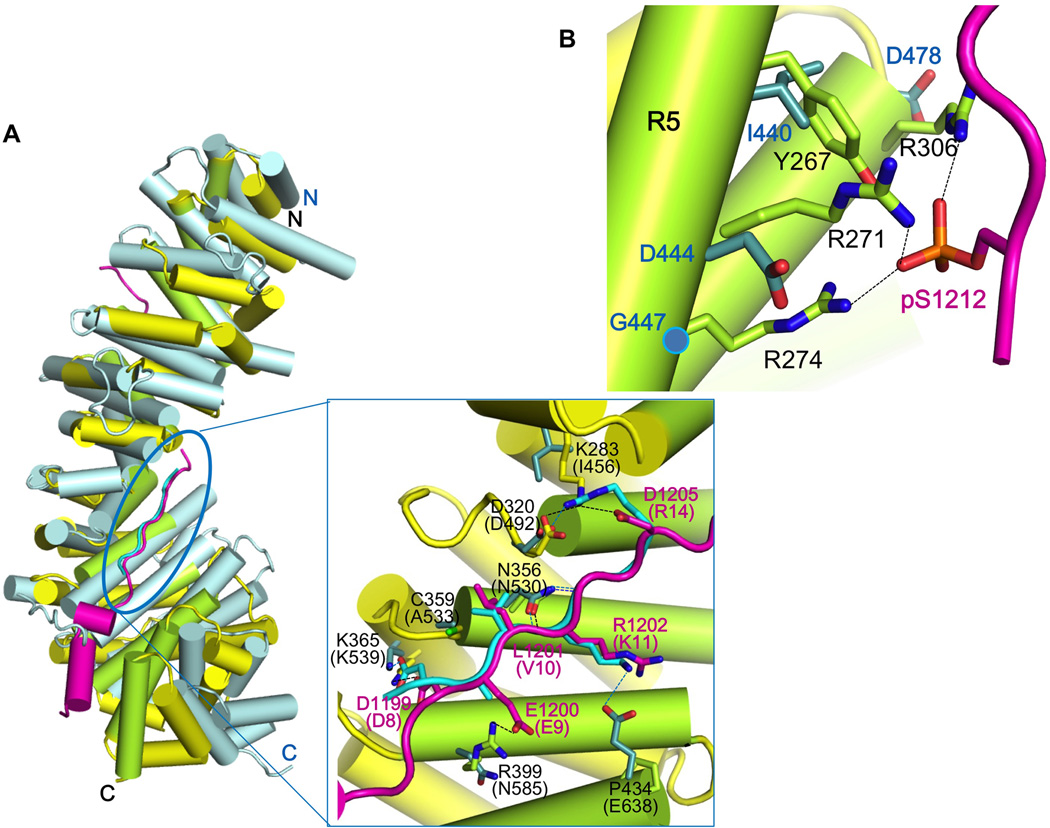
Figure 7. Structural comparison of HMP-2/pHMR-1cyto80 complex with SYS-1/POP-1 complex suggests why only HMP-2 can bind pHMR-1.
(A) Superposition of SYS-1/POP-1 (PDB ID 3C2G) into HMP-2/pHMR-1cyto80 complex. SYS-1 and POP-1 are colored light blue and cyan, respectively. Repeats 7 to 9 of SYS-1 and POP-1 are manually aligned to corresponding regions of HMP-2 and pHMR-1 in Coot. POP-1 (amino acids 8–14) aligns well with pHMR-1 region III, as shown in the close-up (boxed) view. SYS-1 and POP-1 residue numbers are in parentheses, and only those SYS-1 side chains that interact with POP-1 interaction are shown. Side chain of Glu9 of POP-1 is not modeled in the original structure. Hydrogen bonds in HMP-2/pHMR-1cyto80 and SYS-1/POP-1 complexes are shown as black and blue dotted lines, respectively.
(B) Repeats 5 and 6 of SYS-1 are aligned to corresponding region of HMP-2. Three arginine residues of HMP-2 interacting with pS1212 are not conserved in SYS-1.