Figure 2.
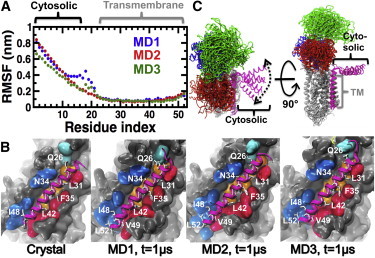
Structural characteristics of PLB in the complex. (A) Cα RMSFs of PLB calculated from each independent MD trajectory. Brackets show the location of the cytosolic and TM helices of PLB. (B) Interactions between the TM domains of SERCA (surface representation) and PLB (cartoon representation) in the crystal and MD structures. PLB is colored in magenta. Residues of PLB that are important for inhibition are shown as white sticks. These residues are located at the SERCA-PLB interface. TM helices of SERCA that are directly involved PLB binding (TM2 (blue), TM4 (cyan), TM6 (orange), and TM9 (red)) are shown. (C) Cartoon representation of the structural ensembles of SERCA-PLB averaged over all three MD trajectories. SERCA and PLB are color-coded as indicated in Fig. 1. To see this figure in color, go online.
