Figure 1.
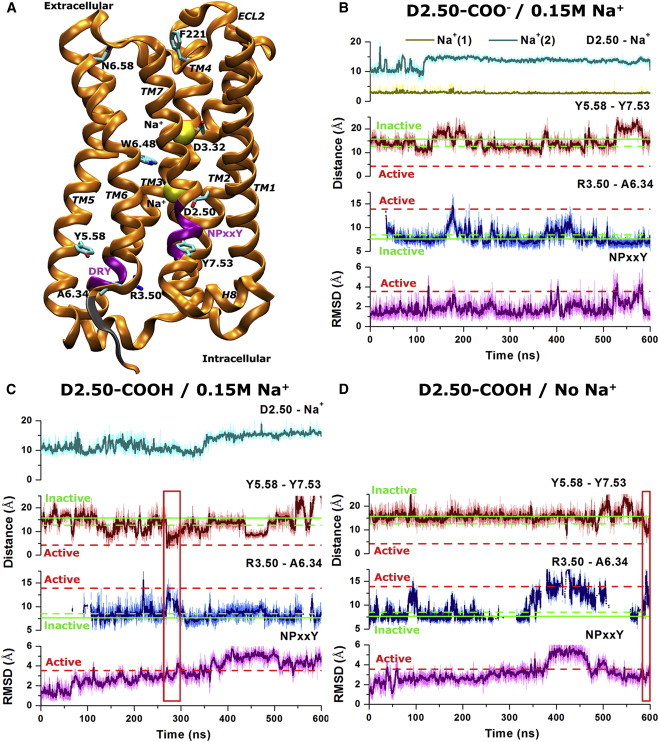
Activation and mechanistic effects of sodium ion binding of the M3 muscarinic receptor simulated via aMD. (A) Schematic representation of the M3 receptor (orange ribbons) with two sodium ions (yellow spheres) bound to the D3.32 orthosteric and D2.50 allosteric sites. Key residues, including D2.50, D3.32, R3.50, A6.34, W6.48, N6.58, Y5.58, Y7.53, and F221ECL2, are shown in sticks, and the two highly conserved DRY and NPxxY motifs are highlighted in purple. Three residues that are missing in the x-ray structure (K6.31−A6.33) are added and shown in gray. (B) In the D2.50-deprotonated receptor in 0.15 M Na+ solution, two sodium ions bind to the D2.50 allosteric and D3.32 orthosteric sites, respectively. (C) In the D2.50-protonated receptor in 0.15 M Na+ solution, only one sodium binds to the D3.32 orthosteric site. (D) A third system with D2.50 protonated is simulated in the absence of Na+. The distances of bound sodium ion(s) to the Cγ atom of D2.50 are plotted in (B) and (C). The corresponding distances between Y5.58 and Y7.53 (hydroxyl oxygens) and between R3.50 and A6.34 (Cα atoms) are plotted for each of the three simulated systems. Thick lines depict the running average over 1 ns (simulation frames were saved every 10 ps for the original). The Y5.58-Y7.53 and R3.50-A6.34 distances in the inactive x-ray structure of the M3 receptor (PDB: 4DAJ) are plotted in the solid green line and those of the inactive and active structures of the M2 receptor (PDB: 3UON and 4MQS, respectively) are plotted in dashed green and red lines, respectively. The R3.50-A6.34 distance is reported only when helical structure is formed up to residue A6.34 in the TM6 intracellular region (Fig. S3). The RMSD of the NPxxY motif between the active and inactive structures of the M2 receptor (3.6 Å) is also plotted in a dashed red line. Red rectangles highlight activation events of the M3 receptor in (C) and (D). To see this figure in color, go online.
