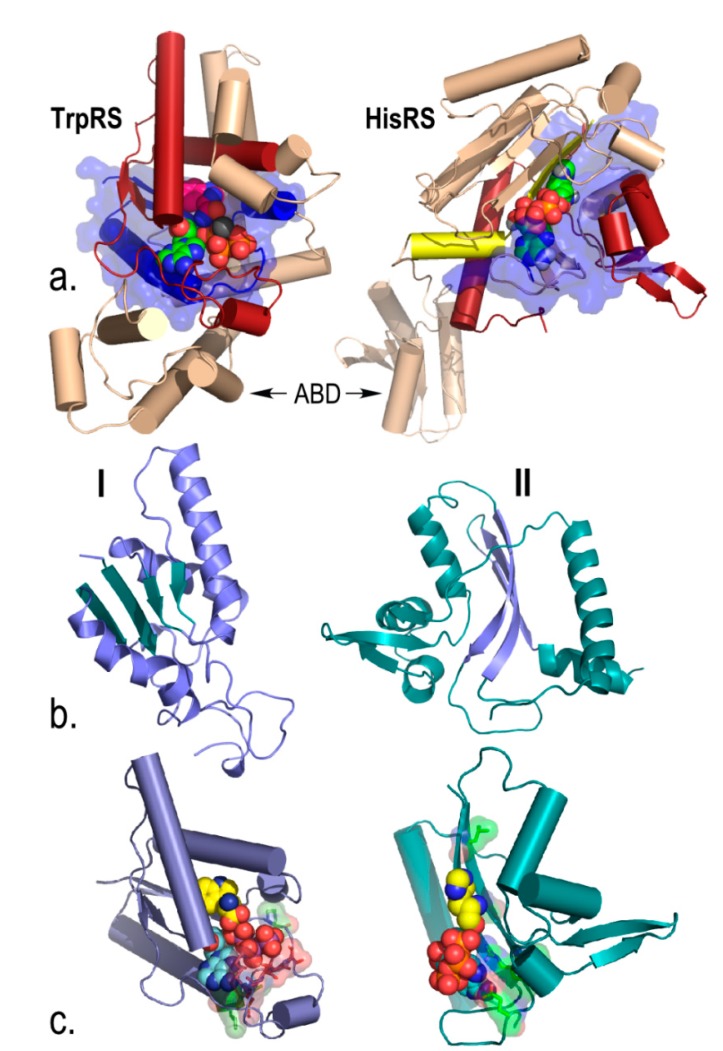
Figure 3.
Urzymes isolated from Class I TrpRS (130 amino acids) and Class II HisRS (124 amino acids). (a) Monomer architectures. Both enzymes are dimeric. Monomers consist of two consensus domains, catalytic and anticodon-binding (ABD). The 46-amino acid ATP binding sites are blue and bounded by transparent surfaces; the remainder of the Urzymes are red. Catalytic domains of both also include insertions, colored amber. Active-site ligands are shown as spheres; (b) Secondary structures are dissimilar; Class I is a Rossmann fold with parallel β-strands; Class II is an antiparallel structure; (c) Amino acid (yellow) and ATP (cyan) substrates are spheres. Stick models of Class I-defining signatures PxxxxHIGH (green) and KMSKS (red) and Class II Motif 2 (green) are surrounded by transparent surfaces to reveal catalytically important interactions with ATP. The cartoons are based on crystal structures of the full-length enzymes. However, long-time MD simulations of both Urzymes in the presence of both substrates have shown that the structures shown here persist, but that in the absence of tryptophan the Class I specificity helix above the bound tryptophan re-orients, removing several key interactions involved in specific recognition [21,25,26].