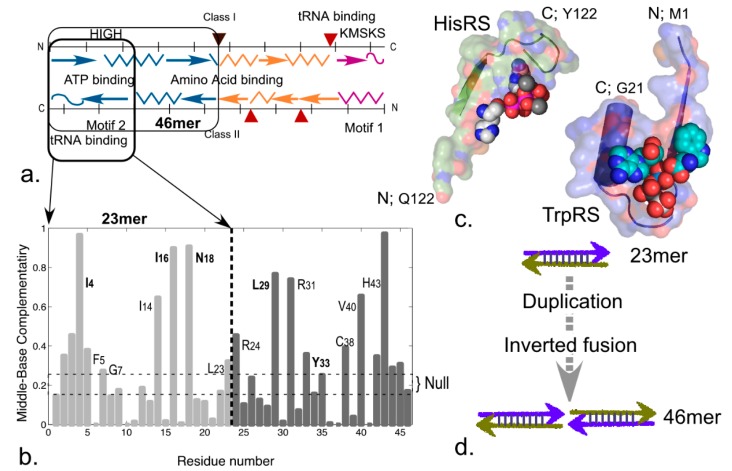
Figure 7.
Evidence for an inverted repeat in coding for Class I ATP binding sites. (a) Schematic of functional divisions in the Class I and II Urzymes, showing parallel localization of ATP, amino acid, and tRNA binding sites; (b) Coding sequences for the 46 residue segment have significantly elevated middle-base pairing (0.28 ± 0.0005 vs. 0.25 ± 0.0004) when aligned antisense to each other. Frequencies of middle-base pairing between residues 4 and 43 together with 16, 18 and 29, 31 and other pairwise comparisons along the gene are evidence of coding by an ancestral palindrome; (c) Structures corresponding to the sense/antisense 23mer containing Class I PxxxHIGH and Class II Motif 2 binding determinants; (d) Putative evolution of the 46mer gene via formation of an inverted repeat. Such a gene would have been stable either as a duplex or as an RNA hairpin.