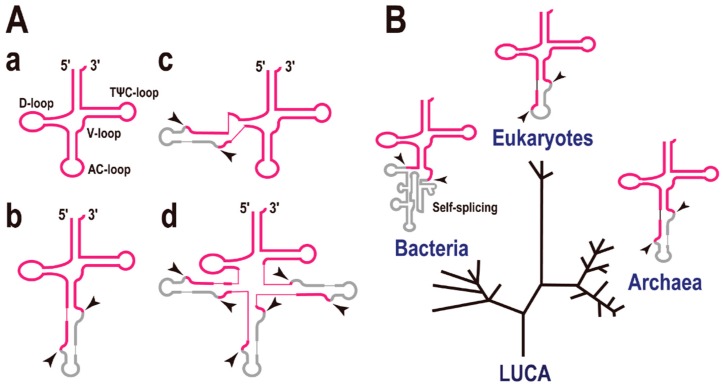
Figure 1.
Intron-containing transfer RNAs (tRNAs). (A) Molecular structures of disrupted tRNAs. Exons are indicated in red and introns in grey. (a) Common tRNA. A description of each stem and loop is included; (b) tRNA containing a single intron at the canonical position, 37/38; (c) An example of a tRNA containing a single intron at a non-canonical position; (d) Example of a tRNA containing multiple introns (up to three introns) at various positions. The two arrowheads indicate the exon-intron boundaries cleaved by the splicing endonuclease. (B) The three domains of life and tRNAs containing a single intron. A phylogenetic tree representing the separation of the Bacteria, Eukaryotes, and Archaea. LUCA, last universal common ancestor. Most introns are located in the anticodon loop region of the tRNAs. Note that the secondary structures of the eukaryotic and archaeal tRNA introns are very similar and that the introns of both are processed by a specific enzyme, the tRNA splicing endonuclease. By contrast, the bacterial tRNA intron is a self-splicing-type intron.