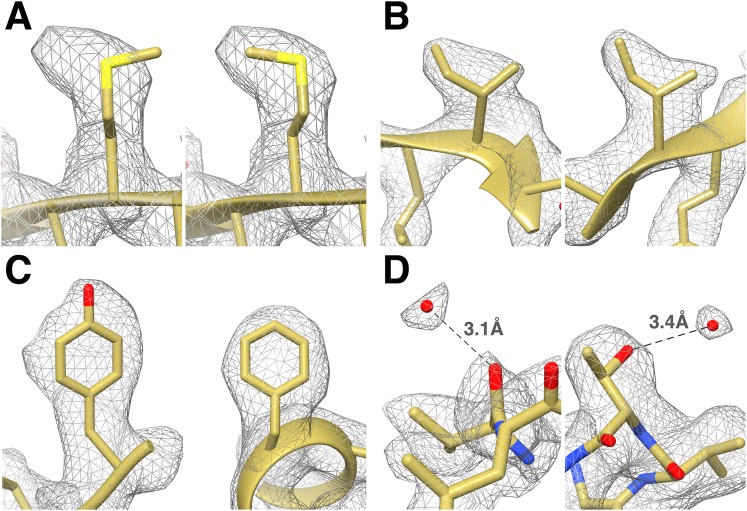
Figure 4. Identification of the rotameric conformation of amino acid side chains and resolving of ordered water molecules in the T20S cryoEM reconstruction at 2.8 Å resolution.
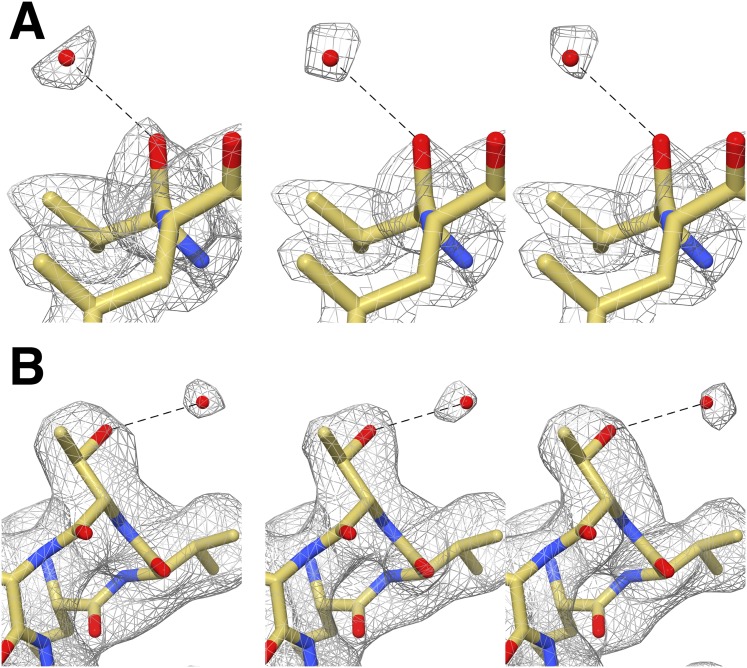
(A) Different rotameric conformations adopted by the Met-14 side chain (β-subunit) between a crystal structure of the T20S determined at 3.4 Å resolution (left, PDB 1PMA) and the EM density (right). (B) Unambiguous establishment of the rotameric conformations of two different isoleucine residues: Ile70 (left, α-subunit) and Ile37 (right, β-subunit). (C) The additional density accounting for the hydroxyl group of tyrosine side chains (left, Tyr132, α-subunit) is prominent when compared to phenylalanine side chains (right, Phe91, α-subunit). (D) Numerous water molecules are resolved in the T20S cryoEM map. Left: a water molecule hydrogen-bonded to the carbonyl group of Val87 (α-subunit). Right: a water molecule hydrogen-bonded to the side chain hydroxyl of Thr102 (α-subunit). The cross-validation of the assignment of these water molecules using gold-standard half maps can be found in Figure 4—figure supplement 1.