Abstract
The parasitoid wasp Nasonia represents a genus of four species that is emerging as a powerful genetic model system that has made and will continue to make important contributions to our understanding of evolutionary biology, development, ecology, and behavior. Particularly powerful are the haplodiploid genetics of the system, which allow some of the advantages of microbial genetics to be applied to a complex multicellular eukaryote. In addition, fertile, viable hybrids can be made among the four species in the genus. This makes Nasonia exceptionally well suited for evolutionary genetics approaches, especially when combined with its haploid genetics and tractability in the laboratory. These features are complemented by an expanding array of genomic, transcriptomic, and functional resources, the application of which has already made Nasonia an important model system in such emerging fields as evolutionary developmental biology and microbiomics. This article describes the genetic and genomic advantages of Nasonia wasps and the resources available for their genetic analysis.
Keywords: Nasonia, genomics, haplodiploid, hybrid genetics, model organisms
NASONIA VITRIPENNIS has been a model for genetic analysis for more than half a century (Whiting 1967), and the past decade has seen a rapid increase in the number and power of its genetic toolkit. These tools, in combination with the wealth of interesting biology (Werren et al. 2010), make the Nasonia system one of the premier genetic systems among insects.
Life History of Nasonia Wasps
Nasonia wasps are parasitoids, meaning that they use a host for nourishment for only the larval portion of their life cycle, and the host is killed in this process (Whiting 1967). N. vitripennis is the most commonly used laboratory model system in the genus and is distributed worldwide in association with human populations (Whiting 1967). Three additional species in the Nasonia genus have been described that have much more restricted distributions in North America. N. giraulti and N. oneida are found in the Northeast, and N. longicornis is restricted to the Northwest (Darling and Werren 1990; Raychoudhury et al. 2010).
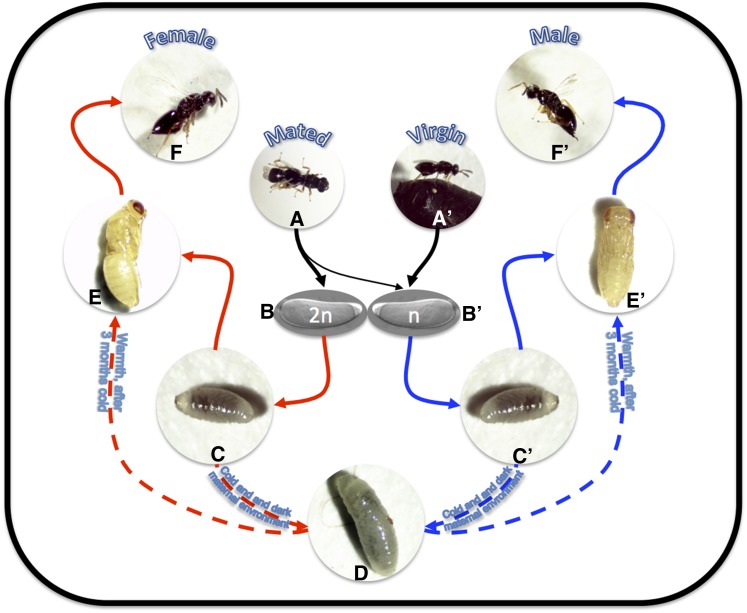
These wasps develop fairly rapidly, with a generation time of 14 days at 25° or 10 days at 28°. Embryogenesis is also quite rapid in these wasps, with larvae emerging a little more than 24 hr after egg laying (Pultz et al. 2005). After larval emergence, the wasps begin to feed on the host hemolymph and undergo up to four larval instars before the onset of pupation (Pultz and Leaf 2003) (Figure 1).
Figure 1.
Life cycle of Nasonia wasps. Both mated (A) and virgin (A′) females will readily lay eggs. Mated females (B) will produce predominantly (80–90%) fertilized eggs, while virgins (B′) can only lay unfertilized eggs. (C and C′) Embryos will develop into legless, simplified larvae. (D) If the mother experienced cold and short-daylight environments, she is more likely to program her offspring to enter diapause, which occurs in the last larval instar. Diapause larvae can survive for more than a year and require an extended cold period to progress in development. (E and E’) As holometabolous insects, Nasonia undergo an extended pupal stage. (F and F’) Adults deriving from fertilized eggs will normally be diploid females, while those from unfertilized eggs will be haploid males.
Like all Hymenoptera, Nasonia use haplodiploid sex determination, which means that unfertilized eggs give rise to haploid males, while fertilized eggs give rise to diploid females (Whiting 1967) (Figure 1). The existence of haploid males is one of the most important features of these wasps, and the many ways that this haploidy can be exploited experimentally will be described later.
It should be noted here that although haplodiploid sex determination is ancestral and highly conserved among the Hymenoptera, the molecular basis for this process is variable. One mode, employed by honeybees and many others, is complementary sex determination (CSD) (Whiting 1943). In this mode, the presence or absence of heterozygosity at a sex-determining locus leads to female or male development, respectively. Normally, there is sufficient polymorphism in an outbred population to prevent the mating of individuals carrying the same allele, so fertilized eggs will develop as heterozygous diploid females. Unfertilized eggs are hemizygous but are effectively homozygous and develop as males. If inbreeding occurs, there is the possibility that a male and a female carry the same allele, and half their progeny will be nonviable diploid males (Whiting 1943).
It is clear that Nasonia do not use a CSD system (Dobson and Tanouye 1998). Nasonia can be heavily inbred (i.e., brother-sister and mother-son matings) for many generations without the appearance of diploid males or any other observable defects (Skinner and Werren 1980; Dobson and Tanouye 1998). Therefore, a single-locus CSD system is excluded, and a multilocus CSD system is still highly unlikely. The currently favored hypothesis is that sex determination relies on differential imprinting on one or more loci in the genomes of the gametes (Beukeboom and van de Zande 2010; Verhulst et al. 2010). This hypothesis rests on dependence of “femaleness” on the presence of a functioning male chromosome set during early embryogenesis. A locus that can confer “femaleness” may be imprinted and repressed on the maternally derived chromosome but is unimprinted on the male chromosomes. Thus, only when the male genome is able to participate in development is the female form generated. The nature of the imprint and the identity of the locus imprinted are as yet unknown (Zwier et al. 2012; Verhulst et al. 2013).
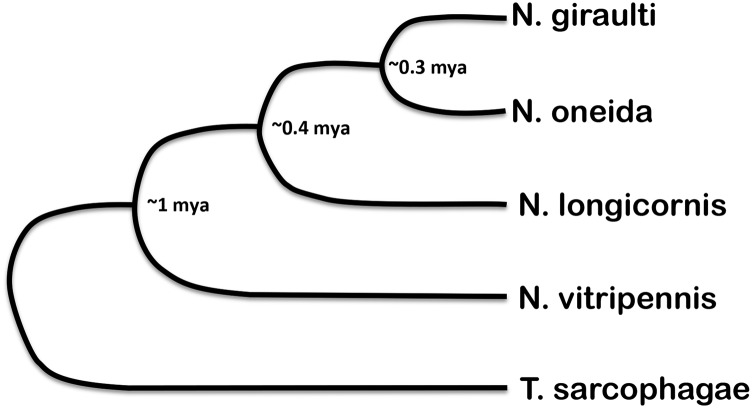
Another noteworthy feature of Nasonia is the ability to make fertile, viable hybrids between any of the four species by curing them of otherwise incompatible Wolbachia infections (Breeuwer and Werren 1990). N. vitripennis is about 1 million years diverged from the other three species, while the latter are from 300,000 to 400,000 years diverged from one another (Werren and Loehlin 2009a) (Figure 2). This feature, in combination with all the other advantages of the system, makes Nasonia a premier model organism for genetic dissection of the evolutionary process that has taken place over relatively short periods of time (Werren and Loehlin 2009a).
Figure 2.
Phylogenetic relationships and approximated divergence times among Nasonia species. Adapted from Werren and Loehlin (2009a) and Werren et al. (2010).
Nasonia as a Genetic Model in the Laboratory
Nasonia are very amenable to culture in the laboratory, with hosts that are easily reared in the laboratory (Werren and Loehlin 2009b), and they are also commercially available (Sarcophaga pupae, Carolina Biological Supply Company, Burlington, NC). They are perfectly happy in Drosophila culture tubes or smaller vessels such as plastic test tubes. Recently, a sterile, host-free in vitro method for rearing Nasonia from egg to adult was developed (Brucker and Bordenstein 2012). This technique has already allowed crucial insights into how the wasp interacts with its symbiotic and commensal bacterial communities (Brucker and Bordenstein 2013). More detailed descriptions of how Nasonia can be maintained in the laboratory can be found elsewhere (Werren and Loehlin 2009a), and here only the most useful characteristics of these wasps for genetic experiments will be described.
A major practical advantage of Nasonia biology is the cold hardiness of these wasps at several life stages. Further development or maturation of the wasps can be suspended by incubation at 4° and then reinitiated if they are returned to higher temperatures within a certain time frame. The amenable stages include the embryo (up to 48 hr; J. A. Lynch, personal observation), the early (yellow) pupal stage (2–3 months), the late, fully pigmented pupa (1 month), and the adult wasp (∼3 weeks). In addition, a facultative larval diapause can be induced, which can allow wasp lines to persist for a year or more with no maintenance effort (Figure 1).
The pupae of Nasonia show the body structure of the adult very clearly and are covered with only a thin cuticle (Figure 1). This allows very easy sexing and phenotyping of pupae. These features are quite convenient for setting up genetic crosses because selection of virgin females and mutants can be done at the investigator’s leisure. In addition, the sex ratio can be manipulated in Nasonia to a large degree, taking advantage of haplodiploidy. All-male broods can be easily generated by allowing virgin females to parasitize hosts, while broods of up to 90% female progeny can be generated if mated females are hosted under optimal conditions of low density (Whiting 1967).
In addition to these investigatory conveniences, Nasonia can be used to address a wide variety of biological questions. Not only are they useful as a comparative system for more established models (i.e., Drosophila), but they also have features that cannot be addressed in other systems. For example, Nasonia use DNA methylation for genomic regulation, unlike the model insect Drosophila melanogaster. Approximately a third of genes are methylated in Nasonia, and methylation occurs over gene bodies and is correlated with broad gene expression across development (Wang et al. 2013).
As alluded to earlier, Nasonia are emerging as a uniquely powerful model system for understanding both the nature and evolution of the hologenome (the genetics of how species and their associated symbiotic and commensal microorganisms interact with each other and their environment), and many of the functional tools described later in this paper can be applied to understanding the evolutionary impact of Nasonia’s microbiomic milieu (Bordenstein and Bordenstein 2011; Kent et al. 2011; Brucker and Bordenstein 2013). These tools also include the cellular and developmental genetics of pathologic interactions between Nasonia, its symbionts, and selfish genetic elements (Tram and Sullivan 2000; Ferree et al. 2008; Swim et al. 2012; Akbari et al. 2013). In addition, Nasonia constitute an ideal model for understanding how parasitoids manipulate their hosts using a potent cocktail of biologically active molecules contained in venom (Danneels et al. 2010; De Graaf et al. 2010; Martinson et al. 2014).
Finally, Nasonia have a great potential as a model to understand the evolution of behavior. Each species has a distinct, complex, and quantifiable courtship behavior that could be genetically dissected (Beukeboom and van den Assem 2001). Other examples include understanding the genetics of host preference (Desjardins et al. 2010), sex-ratio decisions (Pannebakker et al. 2011), memory formation and retention (Hoedjes et al. 2012; Hoedjes and Smid 2014; Hoedjes et al. 2014), and circadian rhythms (Paolucci et al. 2013; Bertossa et al. 2014).
Further descriptions of the interest of Nasonia biology can be found elsewhere (Beukeboom and Desplan 2003; Werren et al. 2010) and are outside the scope of this paper. Instead, I will focus on the tools available to leverage the inherent advantages of the Nasonia system.
Nasonia Genomics
Given the unique advantages of Nasonia genetics and the large number of questions that can be addressed with these wasps, the three described (at the time) species of the Nasonia genus were chosen for whole-genome sequencing. N. vitripennis was sequenced and assembled with 6× Sanger shotgun coverage, while N. giraulti and N. longicornis were sequenced to 1× with Sanger chemistry and to 12× with Illumina short reads (Werren et al. 2010) (Table 1).
Table 1. Genomics Resources Available or in Development for Nasonia.
| Resource | Details | References |
|---|---|---|
| Genome sequence N. vitripennis | 6× coverage Sanger array-aided assembly | Werren et al. 2010, Niehuis et al. 2010 |
| Genome sequence N. giraulti | 1× coverage Sanger, 12× short-read Illumina, 10× PacBio, 120× Illumina paired end | Werren et al. 2010, J. A. Lynch, Y. Kelkar, and J. Werren, in preparation |
| Genome sequence N. longicornis | 1× coverage Sanger, 12× short-read Illumina in-progress hybrid assembly | Werren et al. 2010, J. Werren, personal communication |
| Genome sequence N. oneida | In-progress hybrid assembly | J. Werren, personal communication |
| Genome sequence T. sarcophaga | In-progress hybrid assembly | J. Werren, personal communication |
| Curated transcriptomes N. vitripennis | 1.2, 2.0 | http://nasoniabase.org |
| http://arthropods.eugenes.org/EvidentialGene/ | ||
| Genome database | Genome browser, BLAST, data downloads, and more | www.nasoniabase.org |
| RNAi target design, predicted methylation regions | Database of methylated and transcribed regions plus tool for dsRNA design | http://www.waspatlas.com/home |
The ability to make interspecies crosses was used to map contigs and scaffolds to chromosomes, which further improved the assembly of the N. vitripennis genome (Niehuis et al. 2010; Desjardins et al. 2013) (Table 1) and made it one of only a handful of species whose scaffolds have been mapped to chromosomes.
Currently, efforts are underway to improve the coverage and assembly of the other Nasonia species. For example, N. giraulti is being sequenced using paired-end Illumina reads in combination with long Pacific Bioscience single-molecule real-time (SMRT) reads (Table 1). In addition, N. longicornis, the recently described N. oneida, and Trichomalopsis sarcophagae, a representative of the sister genus to Nasonia (Figure 2), are being sequenced using primarily Illumina technology (Table 1). This will be especially useful in determining the polarity of evolutionary change within the Nasonia genus.
In concert with sequencing of the genome, major efforts to characterize and annotate the transcribed portion of the genome were undertaken (Werren et al. 2010). A combined approach of using ESTs and in silico predictions led to the production of a transcriptome annotation that included more than 24,000 genes (Gilbert 2012) (Table 1).
The sequencing of the three Nasonia genomes and transcriptomes allowed the production of high-resolution microarray resources, including tiling arrays covering the entire genome of Nasonia (Wang et al. 2013) and mapping arrays that allow the mapping of interspecies traits (Desjardins et al. 2013). Having well-annotated transcriptomes and genomes also is a great aid in applying analytical tools that take advantage of next-generation sequencing techniques, for example, in using the cufflinks package for the analysis of differential expression in different developmental stages (Trapnell et al. 2012) or in RNA interference (RNAi) cases (Akbari et al. 2013; Sackton et al. 2013).
The Nasonia genome is hosted as part of the Hymenoptera Genome Database project (Munoz-Torres et al. 2011) (Table 1). Here the annotated genome can be browsed using JBrowse and searched with BLAST to identify genes of interest. Other online resources include a genome-wide methylation database and a tool for designing robustly active and specific double-stranded DNA (dsRNA) target sequences for RNAi experiments hosted by the Tauber Lab (Table 1).
Functional Resources
Reverse genetics
The ability to apply RNAi to knockdown gene function in Nasonia was a major factor in its success so far as an emerging model organism, especially in regard to developmental questions. The original method was parental RNAi, where female pupae were injected with dsRNA against a gene of interest, and then phenotypes were evaluated in the resulting progeny (Lynch and Desplan 2006). The technique has been adapted recently to other life stages, including adults, larvae, and embryos (Abdel-Latief et al. 2008; Werren et al. 2009; Rosenberg et al. 2014), greatly expanding the number of questions that can be addressed in Nasonia (Table 2). RNAi has been used to identify novel systems for embryonic axial patterning (Lynch et al. 2006; Ozuak et al. 2014), to uncover the logic of Nasonia sex determination (Verhulst et al. 2010), and to confirm the role of a species-specific enzyme in the production of a novel pheromone component (Niehuis et al. 2013).
Table 2. Reverse Genetic Approaches Available for Nasonia.
| Approach | Works in Nasonia? | References |
|---|---|---|
| RNA interference | Yes (parental, embryonic, larval, adult) | Lynch and Desplan 2006; Abdel-Latief et al. 2008; Werren et al. 2009; Rosenberg et al. 2014 |
| Morpholinos | Yes (embryonic) | Rosenberg et al. 2014 |
| Germ-line transformation | Limited success | C. Desplan, personal communication |
| CRISPR/Cas9 | Preliminary success | J. A. Lynch, personal observation |
| TALENs | Yes | C. Desplan, personal communication |
| Zinc finger nucleases | Not tested | n/a |
Other methods for manipulating candidate gene function have been or are in the process of being developed. Morpholinos have been employed for knocking down gene function in embryos (Rosenberg et al. 2014), which will be especially useful in cases where RNAi has proven to be difficult. In addition, new techniques for genome editing, such as transcription activator-like effector nucleases (TALENs), zinc finger nucleases, and clustered regularly interspersed palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) (Gaj et al. 2013), are in principle applicable to Nasonia given that methods for raising injected embryos to adulthood now have been firmly established in the wasp (Rosenberg et al. 2014) (Table 2).
Forward genetics
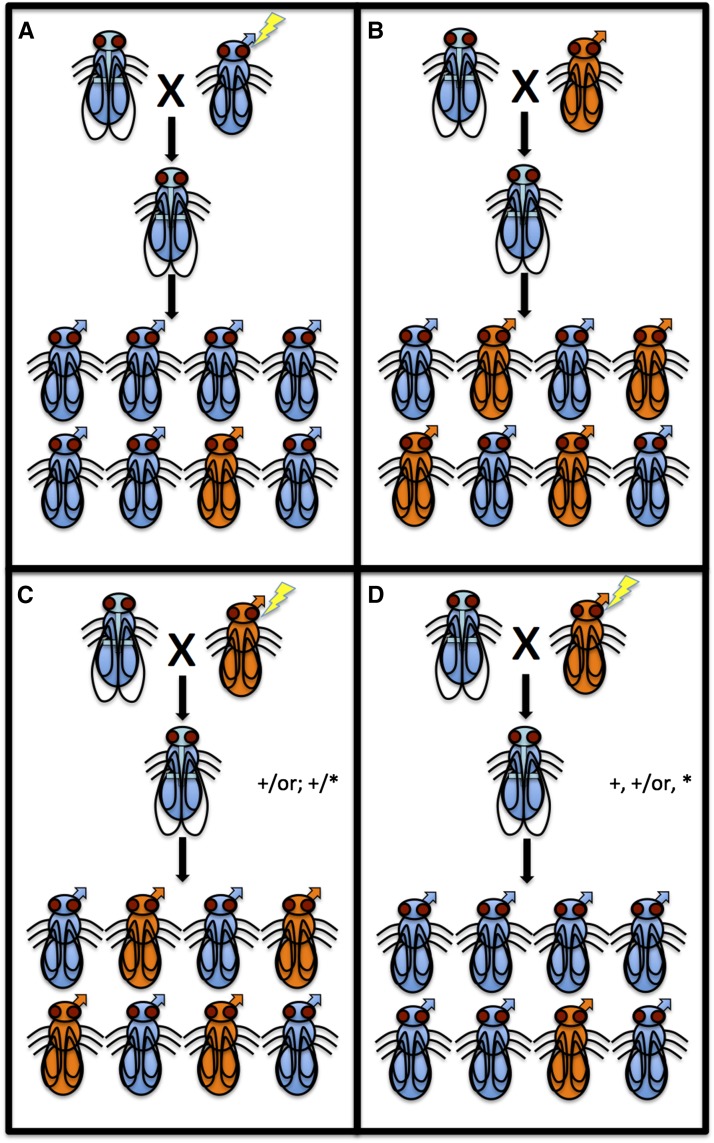
The amenability of Nasonia to classical forward genetics techniques is one of the unique strengths of the system in comparison with other emerging model systems. Nasonia have a small number of chromosomes (five) and the advantage of haploid males, which greatly aid in screening for recessive mutations because phenotypes can be screened in the male progeny of F1 females (Figure 3A). The advantages of haploidy have already been demonstrated by Mary Anne Pultz, whose screens generated and identified many interesting mutations affecting embryonic patterning (Pultz et al. 1999, 2000, 2005).
Figure 3.
Haplodiploid genetics in Nasonia. Males are indicated by an arrow extending from the head and smaller wings. A hypothetical orange (or) mutation is used throughout the figure. (A) A mutation induced in a germ-line cell of a parental-generation male can be screened in the F2 progeny of the male’s virgin daughters. (B) Behavior of a recessive mutation in the Nasonia genetic system. (C) Behavior of a visible mutation and an induced lethal recessive mutation that is not linked. (D) Behavior of a visible mutation and a lethal recessive mutation that are linked. All nonrecombinants carrying the or mutation die, so only relatively rare recombinant orange offspring are observed in F2.
Resources available in Nasonia to aid in identification of the causative lesion in such screens include visible (eye or body color or morphology) marker lines (at least one per chromosome)(Werren and Loehlin 2009a); molecular markers in the form of RAPDs, RFLPs, AFLPs, and SNPs; and the previously mentioned mapping arrays (Gadau et al. 1999; van Opijnen et al. 2005; Niehuis et al. 2010; Desjardins et al. 2013). In addition, a small number of novel mapping strategies were devised in the genomic era that can be leveraged in Nasonia. These include whole-genome resequencing techniques (Blumenstiel et al. 2009) and multiplexed shotgun genotyping (Andolfatto et al. 2011). These techniques will play an important role in overcoming the limitations of the candidate-gene approach in Nasonia, especially in regard to developmental biological questions.
Another useful feature of the Nasonia system is its high tolerance for inbreeding. Unlike other insects (including other hymenopterans), sibling:sibling and offspring:parent matings produce no ill effects in Nasonia. The ability to easily produce isogenic females in combination with the fact that haploid males produce sperm that are genetically identical allows the production of large populations of females with the same genotype. These “clonal” females can be used to test the role of environmental factors in any given mutant or recombinant phenotype (Velthuis et al. 2005; Pannebakker et al. 2011). Velthuis et al. (2005) used this tool to map loci affecting mating preferences in N. longicornis, while Pannebakker et al. (2011) used clonal recombinant lines to map QTL associated with heritable variation in sex allocation in different N. vitripennis populations.
Forward genetic strategies also can be combined with the ease of generating interspecies hybrids to detect naturally occurring differences between Nasonia species. All the mapping resources described in the preceding paragraph can be applied to these traits. In addition, a particularly powerful method to detect recombination events taking place near any particular locus has been developed in Nasonia. In this method, hybrid males showing a phenotype of interest are mutagenized and then mated with wild-type females. The resulting virgin females produce male progeny that are screened for the presence of lethal mutations linked to the causative gene (assessed by a much lower proportion than expected of males showing the trait of interest). Once linkage of a lethal mutation has been detected, these females are mated with wild-type males. Male progeny of the females resulting from this cross are scored, and the survivors that show the phenotype represent recombinants between the linked lethal and causative genes. The power of this technique can be enhanced by combining it with additional linked visible markers (Figure 3C and D). This approach has been used to map the genomic bases of different aspects of interspecific wing size to the cis-regulatory region of the sex-determining gene doublesex (Loehlin et al. 2010) and to multiple changes in the cis-regulatory region of the growth factor unpaired (Loehlin and Werren 2012). The latter is one of the few cases where the causative loci of a morphologic difference between species have been mapped to such high resolution.
Another strategy for mapping interspecies differences also has been devised using a high-density N. vitripennis microarray that can detect the species origin of segments of DNA in the genome of a hybrid animal (Desjardins et al. 2013) (Table 3). This approach was used to map a locus involved in host preference differences between N. giraulti and N. vitripennis to a relatively small genomic region (Desjardins et al. 2010). This approach also led to the identification of a gene responsible for producing a species-specific evolutionarily relevant pheromone whose role was verified using RNAi (Niehuis et al. 2013).
Table 3. Forward Genetic Approaches Applicable to Nasonia.
| Resource | Details | Example |
|---|---|---|
| Haplodiploidy | More rapid identification of mutations | Pultz et al. 1999; Pultz et al. 2000; Pultz et al. 2005 |
| Interspecies hybrids | Detection of evolutionarily relevant variants | Loehlin and Werren 2012 |
| Linked lethal mapping strategy | Detection of interspecies differences | Loehlin et al. 2010 |
| Visible mutations | Aid in mapping and stock maintenance | Pultz et al. 1999; Pultz et al. 2000 |
| Mapping arrays | Genome assembly detecting recombination in hybrids | Desjardins et al. 2013 |
| SNP maps | Narrowing QTL regions | Niehuis et al. 2013 |
| Mapping population | Highly outbred line for selection and mapping of complex traits | Van De Zande et al. 2014 |
| Segmental introgression lines | Defined genomic fragments from one species in the genomic context of another | Desjardins et al. 2013 |
An additional resource available in Nasonia is a set of segmental introgression lines (SILs) in which fragments of the N. giraulti genome, marked either visibly or genetically, were back-crossed into a N. vitripennis background for over eight generations and then were made homozygous (Table 3). The break points of the introgression have been estimated, and these lines serve as a crucial resource for screening for interspecies differences in any trait of interest (Desjardins et al. 2013).
Recently, a highly outbred line of N. vitripennis was established, and a protocol for maintenance of its genetic diversity was devised. This line will be useful for experimental evolution, mapping of complex traits, and population genetics, among others (Van De Zande et al. 2014) (Table 3).
Tools for Molecularly Characterizing and Interpreting Phenotypes
In addition to tools for generating, maintaining, and mapping mutants, a successful genetic model organism needs tools for visualizing and quantifying the activity of genes and their products. Robust techniques for in situ hybridization to detect spatial patterns of mRNA expression have been developed and applied to both embryos and larvae. In the embryo, multiple transcripts can be detected simultaneously with fluorescence (Ozuak et al. 2014), and a technique to detect nascent transcripts in nuclei has been reported recently (Verhulst et al. 2013). High-throughput probe production and hybridization in 96-well plates has been established for Nasonia embryos (J. A. Lynch, in preparation), and a chamber for mass egg lay called the Waspinator (Buchta et al. 2013) will allow for mass screening of gene expression patterns in the Nasonia embryo, as has been done for Drosophila (Tomancak et al. 2002).
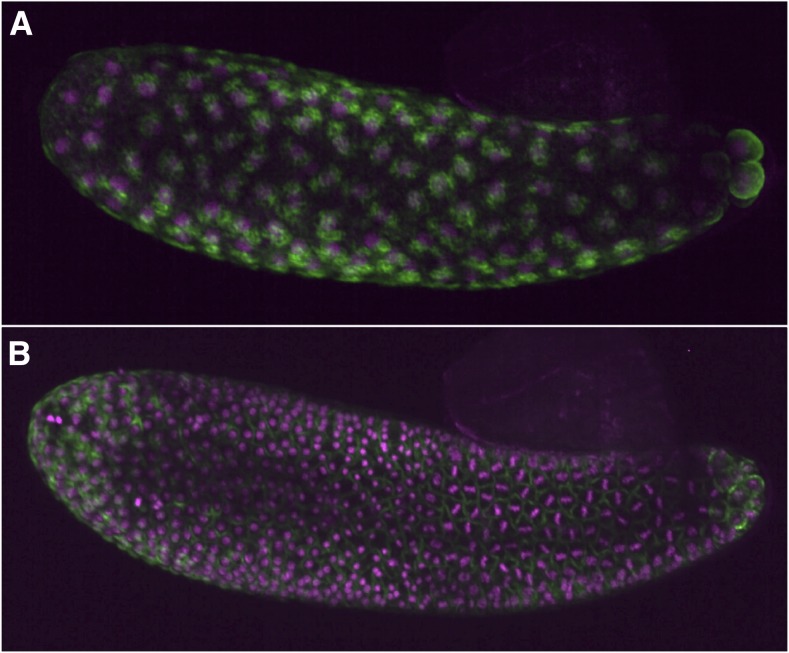
The Nasonia egg is relatively small and optically clear, making it an ideal system for time-lapse analysis of early developmental events. Such analyses have already been performed using differential interference contrast illumination (Buchta et al. 2013) and fluorescent live imaging (J. A. Lynch, personal observation) (Figure 4). The combination of live imaging with the forward and reverse genetic techniques will make Nasonia an even more powerful model for embryonic patterning and morphogenetic processes.
Figure 4.
Live imaging in Nasonia. Nasonia embryos were injected with mRNAs encoding Histone::RFP (magenta, marking chromosomes) and Life Actin::GFP (green, marking f-actin) according to the protocols of Benton et al. (2013). Embryos shown are undergoing the eighth (A) and tenth (B) synchronous syncytial divisions of early embryogenesis. No dechorionation was necessary for injection or microscopy.
The Future
With the increasing power and decreasing cost of next-generation sequencing technologies, the possibility to robustly identify causative nucleotides underlying naturally occurring and laboratory-induced phenotypic differences will increase at an accelerating rate. The Nasonia species group is uniquely well poised to take advantage of these developments. It has a strong foundation of genetic and genomic resources, is the most experimentally tractable haplodiploid genetic model system, and has a still largely untapped promise in its ability to make hybrid and recombinant progeny across species. These possibilities, combined with the evolutionary questions (i.e., venom, development, methylation, hologenomics, biochemistry, population genetics, and speciation, behavior, among many others) that will become increasingly experimentally tractable in the genomic era, make the Nasonia genus one of the most exciting emerging model systems with which to probe the deepest mysteries of biology.
Acknowledgments
I thank three anonymous reviewers for valuable comments and suggestions. I also thank Claude Desplan and Jack Werren for communication of unpublished results, and continuous support throughout the writing of this manuscript.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Abdel-Latief M., Garbe L. A., Koch M., Ruther J., 2008. An epoxide hydrolase involved in the biosynthesis of an insect sex attractant and its use to localize the production site. Proc. Natl. Acad. Sci. USA 105: 8914–8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari O. S., Antoshechkin I., Hay B. A., Ferree P. M., 2013. Transcriptome profiling of Nasonia vitripennis testis reveals novel transcripts expressed from the selfish B chromosome, paternal sex ratio. G3 3: 1597–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfatto P., Davison D., Erezyilmaz D., Hu T. T., Mast J., et al. , 2011. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 21: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M. A., Akam M., Pavlopoulos A., 2013. Cell and tissue dynamics during Tribolium embryogenesis revealed by versatile fluorescence labeling approaches. Development 140: 3210–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertossa R. C., van de Zande L., Beukeboom L. W., Beersma D. G., 2014. Phylogeny and oscillating expression of period and cryptochrome in short and long photoperiods suggest a conserved function in Nasonia vitripennis. Chronobiol. Int. 31: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L., Desplan C., 2003. Nasonia. Curr. Biol. 13: R860. [DOI] [PubMed] [Google Scholar]
- Beukeboom L. W., van den Assem J., 2001. Courtship and mating behaviour of interspecific Nasonia hybrids (Hymenoptera, Pteromalidae): a grandfather effect. Behav. Genet. 31: 167–177. [DOI] [PubMed] [Google Scholar]
- Beukeboom L. W., van de Zande L., 2010. Genetics of sex determination in the haplodiploid wasp Nasonia vitripennis (Hymenoptera: Chalcidoidea). J. Genet. 89: 333–339. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Bordenstein S. R., 2011. Temperature affects the tripartite interactions between bacteriophage WO, Wolbachia, and cytoplasmic incompatibility. PLoS ONE 6: e29106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer J. A. J., Werren J. H., 1990. Microorganisms associated with chromosome destruction and reproductive isolation between two insect species. Nature 346: 558–560. [DOI] [PubMed] [Google Scholar]
- Brucker R. M., Bordenstein S. R., 2012. In vitro cultivation of the hymenoptera genetic model, Nasonia. PLoS ONE 7: e51269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucker R. M., Bordenstein S. R., 2013. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341: 667–669. [DOI] [PubMed] [Google Scholar]
- Buchta T., Ozuak O., Stappert D., Roth S., Lynch J. A., 2013. Patterning the dorsal-ventral axis of the wasp Nasonia vitripennis. Dev. Biol. 381: 189–202. [DOI] [PubMed] [Google Scholar]
- Danneels E. L., Rivers D. B., de Graaf D. C., 2010. Venom proteins of the parasitoid wasp Nasonia vitripennis: recent discovery of an untapped pharmacopee. Toxins 2: 494–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling D. C., Werren J. H., 1990. Biosystematics of Nasonia (Hymenoptera, Pteromalidae): two new species reared from birds’ nests in North America. Ann. Entomol. Soc. Am. 83: 352–370. [Google Scholar]
- de Graaf D. C., Aerts M., Brunain M., Desjardins C. A., Jacobs F. J., et al. , 2010. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 19(Suppl. 1): 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Gadau J., Lopez J. A., Niehuis O., Avery A. R., et al. , 2013. Fine-scale mapping of the Nasonia genome to chromosomes using a high-density genotyping microarray. G3 3: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins C. A., Perfectti F., Bartos J. D., Enders L. S., Werren J. H., 2010. The genetic basis of interspecies host preference differences in the model parasitoid Nasonia. Heredity 104: 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson S. L., Tanouye M. A., 1998. Evidence for a genomic imprinting sex determination mechanism in Nasonia vitripennis (Hymenoptera; Chalcidoidea). Genetics 149: 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Avery A., Azpurua J., Wilkes T., Werren J. H., 2008. A bacterium targets maternally inherited centrosomes to kill males in Nasonia. Curr. Biol. 18: 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau J., Page R. E., Jr, Werren J. H., 1999. Mapping of hybrid incompatibility loci in Nasonia. Genetics 153: 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. G., 2012. Evigene genes for Nasonia vitripennis. Available at: http://arthropods.eugenes.org/EvidentialGene/. Accessed: February 14, 2015.
- Hoedjes K. M., Smid H. M., 2014. Natural variation in long-term memory formation among Nasonia parasitic wasp species. Behav. Processes 105: 40–45. [DOI] [PubMed] [Google Scholar]
- Hoedjes K. M., Smid H. M., Vet L. E., Werren J. H., 2014. Introgression study reveals two quantitative trait loci involved in interspecific variation in memory retention among Nasonia wasp species. Heredity 113: 542–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoedjes K. M., Steidle J. L., Werren J. H., Vet L. E., Smid H. M., 2012. High-throughput olfactory conditioning and memory retention test show variation in Nasonia parasitic wasps. Genes Brain Behav. 11: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent B. N., Funkhouser L. J., Setia S., Bordenstein S. R., 2011. Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS ONE 6: e24984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin D. W., Enders L. S., Werren J. H., 2010. Evolution of sex-specific wing shape at the widerwing locus in four species of Nasonia. Heredity 104: 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin D. W., Werren J. H., 2012. Evolution of shape by multiple regulatory changes to a growth gene. Science 335: 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. A., Brent A. E., Leaf D. S., Pultz M. A., Desplan C., 2006. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439: 728–732. [DOI] [PubMed] [Google Scholar]
- Lynch J. A., Desplan C., 2006. A method for parental RNA interference in the wasp Nasonia vitripennis. Nat. Protoc. 1: 486–494. [DOI] [PubMed] [Google Scholar]
- Martinson E. O., Wheeler D., Wright J., Mrinalini A. L., Siebert, et al. , 2014. Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol. Ecol. 23: 5918–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Torres M. C., Reese J. T., Childers C. P., Bennett A. K., Sundaram J. P., et al. , 2011. Hymenoptera Genome Database: integrated community resources for insect species of the order Hymenoptera. Nucleic Acids Res. 39: D658–D662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehuis O., Buellesbach J., Gibson J. D., Pothmann D., Hanner C., et al. , 2013. Behavioural and genetic analyses of Nasonia shed light on the evolution of sex pheromones. Nature 494: 345–348. [DOI] [PubMed] [Google Scholar]
- Niehuis O., Gibson J. D., Rosenberg M. S., Pannebakker B. A., Koevoets T., et al. , 2010. Recombination and its impact on the genome of the haplodiploid parasitoid wasp Nasonia. PLoS ONE 5: e8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozuak O., Buchta T., Roth S., Lynch J. A., 2014. Dorsoventral polarity of the Nasonia embryo primarily relies on a BMP gradient formed without input from Toll. Curr. Biol. 24: 2393–2398 [DOI] [PubMed] [Google Scholar]
- Pannebakker B. A., Watt R., Knott S. A., West S. A., Shuker D. M., 2011. The quantitative genetic basis of sex ratio variation in Nasonia vitripennis: a QTL study. J. Evol. Biol. 24: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolucci S., van de Zande L., Beukeboom L. W., 2013. Adaptive latitudinal cline of photoperiodic diapause induction in the parasitoid Nasonia vitripennis in Europe. J. Evol. Biol. 26: 705–718. [DOI] [PubMed] [Google Scholar]
- Pultz M. A., Leaf D. S., 2003. The jewel wasp Nasonia: querying the genome with haplo-diploid genetics. Genesis 35: 185–191. [DOI] [PubMed] [Google Scholar]
- Pultz M. A., Pitt J. N., Alto N. M., 1999. Extensive zygotic control of the anteroposterior axis in the wasp Nasonia vitripennis. Development 126: 701–710. [DOI] [PubMed] [Google Scholar]
- Pultz M. A., Westendorf L., Gale S. D., Hawkins K., Lynch J., et al. , 2005. A major role for zygotic hunchback in patterning the Nasonia embryo. Development 132: 3705–3715. [DOI] [PubMed] [Google Scholar]
- Pultz M. A., Zimmerman K. K., Alto N. M., Kaeberlein M., Lange S. K., et al. , 2000. A genetic screen for zygotic embryonic lethal mutations affecting cuticular morphology in the wasp Nasonia vitripennis. Genetics 154: 1213–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhury R., Desjardins C. A., Buellesbach J., Loehlin D. W., Grillenberger B. K., et al. , 2010. Behavioral and genetic characteristics of a new species of Nasonia. Heredity 104: 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. I., Brent A. E., Payre F., Desplan C., 2014. Dual mode of embryonic development is highlighted by expression and function of Nasonia pair-rule genes. eLife 3: e01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton T. B., Werren J. H., Clark A. G., 2013. Characterizing the infection-induced transcriptome of Nasonia vitripennis reveals a preponderance of taxonomically-restricted immune genes. PLoS ONE 8: e83984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner S. W., Werren J. H., 1980. The genetics of sex determination in Nasonia vitripennis (Hymenoptera, Pteromalidae). Genetics 94: s98. [Google Scholar]
- Swim M. M., Kaeding K. E., Ferree P. M., 2012. Impact of a selfish B chromosome on chromatin dynamics and nuclear organization in Nasonia. J. Cell Sci. 125: 5241–5249. [DOI] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., et al. , 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: research0088–0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram U., Sullivan W., 2000. Reciprocal inheritance of centrosomes in the parthenogenetic hymenopteran Nasonia vitripennis. Curr. Biol. 10: 1413–1419. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Zande L., Ferber S., de Haan A., Beukeboom L. W., van Heerwaarden J., et al. , 2014. Development of a Nasonia vitripennis outbred laboratory population for genetic analysis. Mol. Ecol. Resour. 14: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Baudry E., Baldo L., Bartos J., Werren J. H., 2005. Genetic variability in the three genomes of Nasonia: nuclear, mitochondrial and Wolbachia. Insect Mol. Biol. 14: 653–663. [DOI] [PubMed] [Google Scholar]
- Velthuis B. J., Yang W. C., van Opijnen T., Werren J. H., 2005. Genetics of female mate discrimination of heterospecific males in Nasonia (Hymenoptera, Pteromalidae). Anim. Behav. 69: 1107–1120. [Google Scholar]
- Verhulst E. C., Beukeboom L. W., van de Zande L., 2010. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science 328: 620–623. [DOI] [PubMed] [Google Scholar]
- Verhulst E. C., Lynch J. A., Bopp D., Beukeboom L. W., van de Zande L., 2013. A new component of the Nasonia sex determining cascade is maternally silenced and regulates transformer expression. PLoS ONE 8: e63618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wheeler D., Avery A., Rago A., Choi J. H., et al. , 2013. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9: e1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Loehlin D. W., 2009a The parasitoid wasp Nasonia: an emerging model system with haploid male genetics. Cold Spring Harb Protoc. 2009: pdb.emo134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Loehlin D. W., 2009b Rearing Sarcophaga bullata fly hosts for Nasonia (parasitoid wasp). Cold Spring Harb Protoc. 2009: pdb.prot5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Loehlin D. W., Giebel J. D., 2009. Larval RNAi in Nasonia (parasitoid wasp). Cold Spring Harb Protoc. 2009: pdb.prot5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J. H., Richards S., Desjardins C. A., Niehuis O., Gadau J., et al. , 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327: 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting A. R., 1967. The biology of the parasitic wasp Mormoniella vitripennis [=Nasonia brevicornis] (Walker). Q. Rev. Biol. 42: 333–406. [Google Scholar]
- Whiting P. W., 1943. Multiple alleles in complementary sex determination of Habrobracon. Genetics 28: 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwier M. V., Verhulst E. C., Zwahlen R. D., Beukeboom L. W., van de Zande L., 2012. DNA methylation plays a crucial role during early Nasonia development. Insect Mol. Biol. 21: 129–138. [DOI] [PubMed] [Google Scholar]