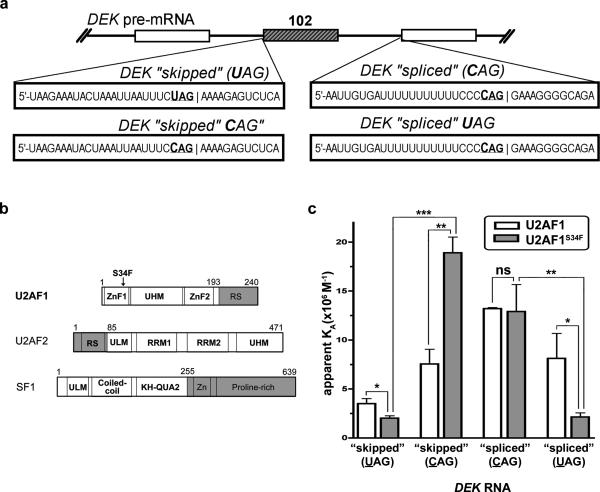
Figure 4. Comparison of the WT and S34F U2AF1 affinities for splice sites from a representative affected transcript, the DEK oncogene.
(a) DEK exon (exon 3) and RNA sequences used in the experiment. (b) Boundaries of recombinant protein constructs used in this study (white). The U2AF2 and SF1 subunits are included. Both U2AF proteins are full- length with the exception of the nonspecific RS domain (gray). KH-QUA2, K-Homology Quaking motif; UHM, U2AF homology motif; ULM, U2AF ligand motif; ZnF, zinc finger. (c) Bar graph of DEK-“skipped”(UAG), DEK-“skipped”-CAG, DEK-“spliced”(CAG), or DEK-“spliced”-UAG RNA affinities of either wild-type U2AF1 (white) or the S34F mutant (gray) in purified U2AF2/SF1- complexes. Average values and standard deviations of three experiments are given. ns, not significant (P>0.3); *P<0.03, **P<0.003, ***P<0.0003.