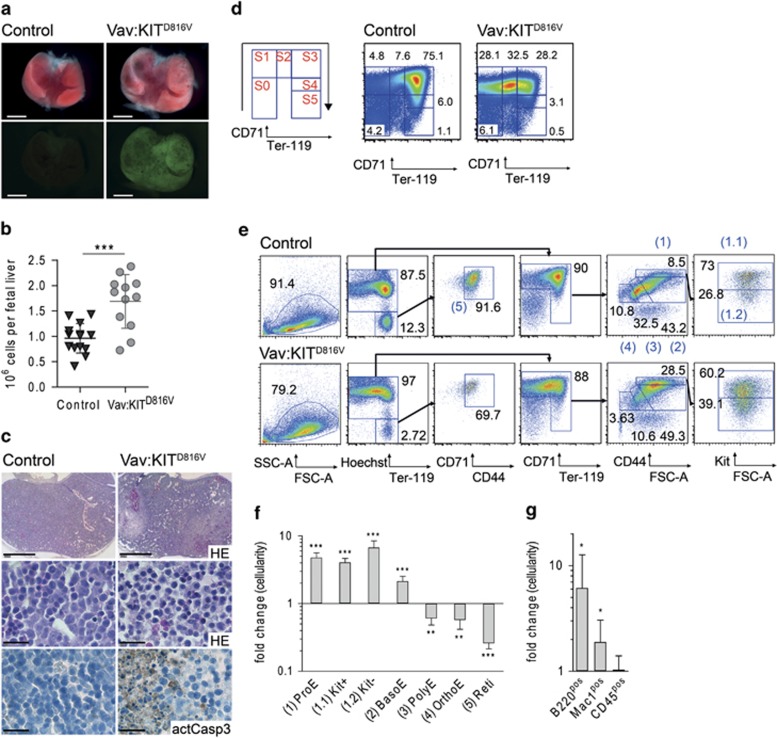
Figure 2.
Accumulation of early erythroblast precursor cells in fetal liver of KITD816V mutant mice. (a) R26-LSL-KITD816V mice were mated to Vav-iCre mice to induce ectopic KITD816V in hematopoietic cells. Pictures show fetal livers from E13.5 control embryos and embryos expressing KITD816V, latter indicated by GFP reporter fluorescence. Scale bar: 1 mm. (b) Cellularity of E13.5 fetal livers. Vav:KITD816V: N=12. Controls: N=14. (c) Morphological appearance of E13.5 fetal liver tissue in hematoxylin and eosin staining of embryo transversal sections in low (top) and high magnification (middle). Immunohistochemical staining of active caspase 3 (bottom). Scale bar: 1 mm (top) and 20 μm (middle, bottom). (d) Staging of E13.5 fetal liver cells based on CD71/Ter119 expression. Mac1negB220neg cells were gated for analysis. (e) Gating strategy for flow cytometric analysis of erythropoiesis beginning from ProE stage. Black numbers indicate cell frequencies in relation to parent gate. Gates numbered in blue define erythroid cells in successive stages of differentiation and are quantified and named in (f). (f and g) Graphs show fold change in total cellularity for Vav:KITD816V animals in relation to controls in log10 scale. Variance is presented as standard deviation. P-values were determined using a two-tailed, unpaired Student's t-test; *P<0.05; **P<0.01; ***P<0.001. (f) Vav:KITD816V: N=4. Controls: N=4. Kit+: Kitpos ProEs; Kit-: Kitneg ProEs; BasoE, basophilic erythroblast; OrthoE, orthochromatic erythroblast; PolyE, polychromatic erythroblast; Reti: reticulocyte. (g) Vav:KITD816V: N=7. Controls: N=9