Fig. 1.
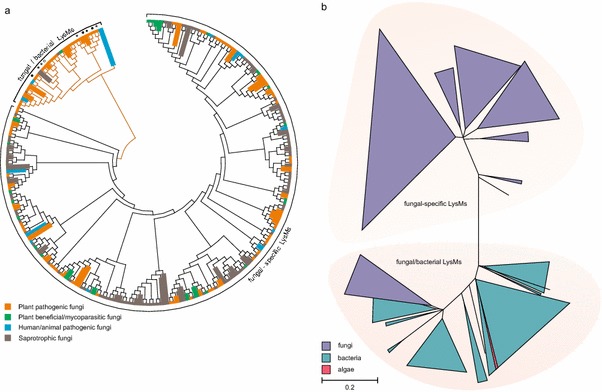
Phylogenetic analysis of fungal LysM motifs. a Phylogenetic analysis of fungal LysMs extracted from the MycoCosm JGI genome portal (Grigoriev et al. 2014). The following fungal genomes were analysed: Neurospora crassa, Aspergillus nidulans, A. fumigatus, A. niger, Trichoderma reesei, T. atroviride, T. virens, Nectria haematococca (Fusarium solani), Mycosphaerella graminicola, Phycomyces blakesleeanus, Cochliobolus heterostrophus, Batrachochytrium dendrobatidis, Phanerochaete chrysosporium, Laccaria bicolor, Postia placenta, Botrytis cinerea, Candida albicans, Fusarium graminearum, F. oxysporum, Magnaporthe grisea, Sclerotinia sclerotiorum and this set was amended with Cladosporium fulvum Ecp6. LysM motifs were aligned using ClustalX (Thompson et al. 1997). Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013) using the Neighbor-Joining method. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Interior-branch test with 1,000 bootstrap replications was performed and only branches with a confidence probability greater than 95 % are shown. Within the fungal/bacterial clade of LysMs, stars indicate the position of the three LysM domains of C. fulvum Ecp6, diamonds correspond to the two Slp1 LysMs of M. grisea and black and white circles indicate the LysMs of Mg3LysM and Mg1LysM of M. graminicola, respectively. b Phylogenetic analysis of LysMs derived from a protein BLAST search using the 5th LysM of Tal6, the 1st LysM of Ecp6 and the 1st LysM of Slp1 as queries. Alignment and evolutionary analysis of LysMs was performed as described for (a)
