Fig. 3.
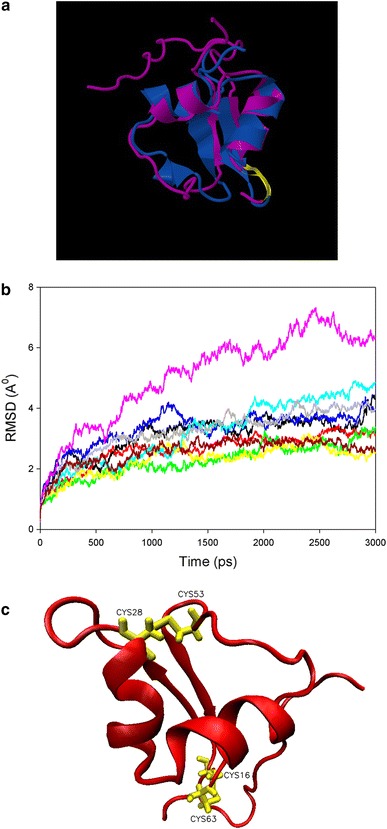
Structural modelling and molecular dynamics simulations of Tal6-5LysM. a Superimposed structures of the homology modelling template MltD (PDB ID:1E0G) (purple) and the modelled structure Tal6-5LysM (pink) with Z-score 3.89, RMSD 2.49 A and sequence similarity of the superimposed region 42 %. Models were constructed using the software Modeller 8v2 (Sali and Blundell 1993). The superimposed structures were created using the jCE algorithm for Combinatorial Extension (http://source.rcsb.org/jfatcatserver/index.jsp). 50 % of the substrate binding loop, between strand1 and helix1, are conserved and this region contains the ligand-binding residue Asp. The predicted substrate binding loop is highlighted in yellow. b RMSD of Cα residues along 3 ns simulations at 300 K. Black line Unpatched model, Red line CYS16–CYS28, Green line CYS16–CYS53, Yellow line CYS16–CYS63, Blue line CYS28–CYS53, Pink line CYS28–CYS63, Cyan line CYS53–CYS63, Grey line CYS16–CYS28:CYS53–CYS63, Dark red line CYS16–CYS63:CYS28–CYS53. c TAL6-5LysM structure at the end of 3 ns MD simulation at 300 K containing two disulphide bridges between Cys16–Cys63 and Cys28–Cys53. Cysteines are indicated in yellow. The numbering of the cysteines corresponds to their position in the amino acid sequence of Tal6-5LysM. For MD simulations, the NAMD/VMD software package was used (Phillips et al. 2005). The initial structure was modified by patching the possible disulphide bridge combinations using psfgen script of the VMD program. The Solvate program of the VMD package and TIP3P water were used for the water box. A 2 fs timestep was used and data collection was performed at every 2 ps. Structures were minimized 50,000 steps initially using conjugate gradient method and then equilibrated for 1 ns at 300 K. MD simulations were performed for each single patch, double patch and unpatched model in 6 A° water box using periodic boundary conditions. Additionally, 9 ns MD simulations for minimized and equilibrated structures of each model were performed at 300 K using the same method. Coordinates of Cys16–Cys28, Cys16–Cys63, Cys53–Cys63 and Cys16–Cys63:Cys28–Cys53 at the end of 9 ns simulations at 300 K were used as initial structures for equilibration steps of MD simulations at 550 K for 1 ns. Then, the equilibrated structures were simulated for an additional 1 ns at 550 K. RMSDs and radius of gyration of each simulation were calculated from the trajectory files using VMD. Total energy of the system along the simulation was extracted for each model from log files generated during the simulations using VMD and moving averages of the simulations were calculated for each model. For 3 ns MD simulations, the averages of three simulations were calculated. RMSD and total energy values for each simulation were compared
