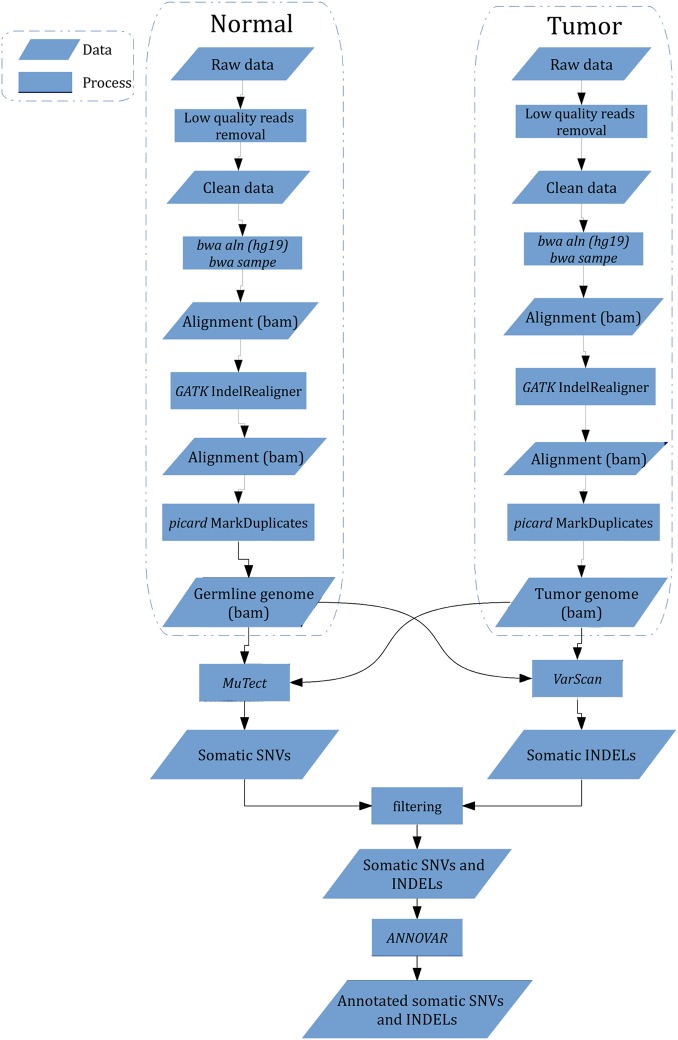
Figure 1.
Work flow of identification of somatic single-nucleotide variation (SNV) and insertion and deletion (INDEL) from raw sequencing data. Low quality reads were removed, and bwa was used to perform alignment followed by alignment calibration by GATK and marked duplicates by picard. MuTect and VarScan were employed to detect somatic SNV and INDEL, processed by further filtering to eliminate false positives, respectively. All mutations were annotated with ANNOVAR.