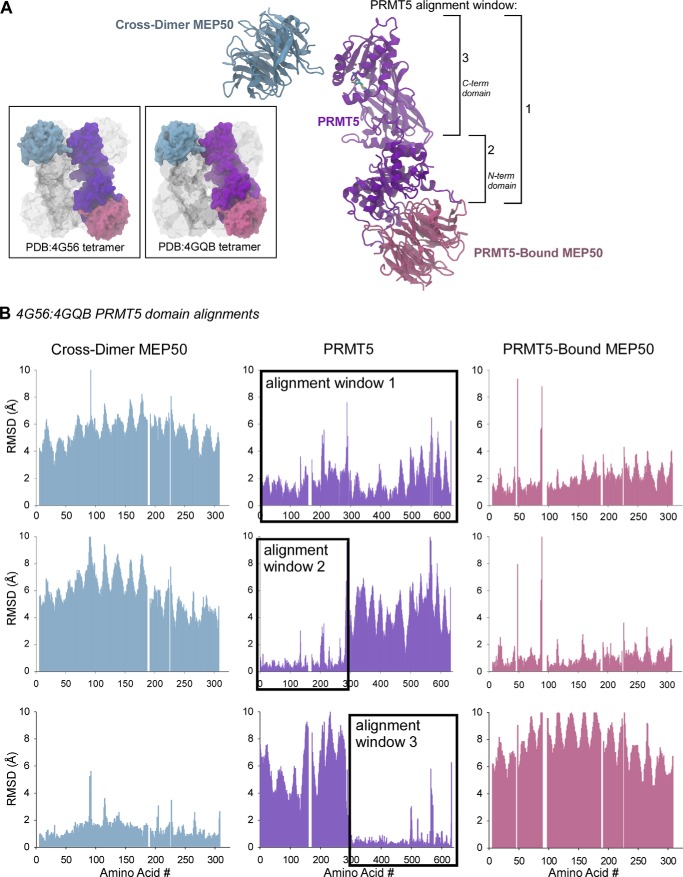
FIGURE 1.
Evolutionarily conserved spatial arrangement between the PRMT5 catalytic domain and its cross-dimer paired MEP50. A, ribbon diagram of the Xenopus PRMT5 monomer (purple), its directly bound MEP50 molecule (pink), and the cross-dimer MEP50 (blue). Structural alignment windows 1, 2, and 3 are indicated on the right. Inset boxes, surface diagrams of the Xenopus (Protein Data Bank code 4G56) and human (Protein Data Bank code 4GQB) PRMT5-MEP50 structures, with the analyzed PRMT5 and MEP50 molecules colored as in A; the remainder of the structures are shown in gray. B, per-residue Cα alignments between the Xenopus and human PRMT5 windows 1, 2, and 3 as calculated with VMD MultiSeq, plotted as RMSD (Å) against amino acid position in the sequence. The PRMT5 alignment windows are boxed.