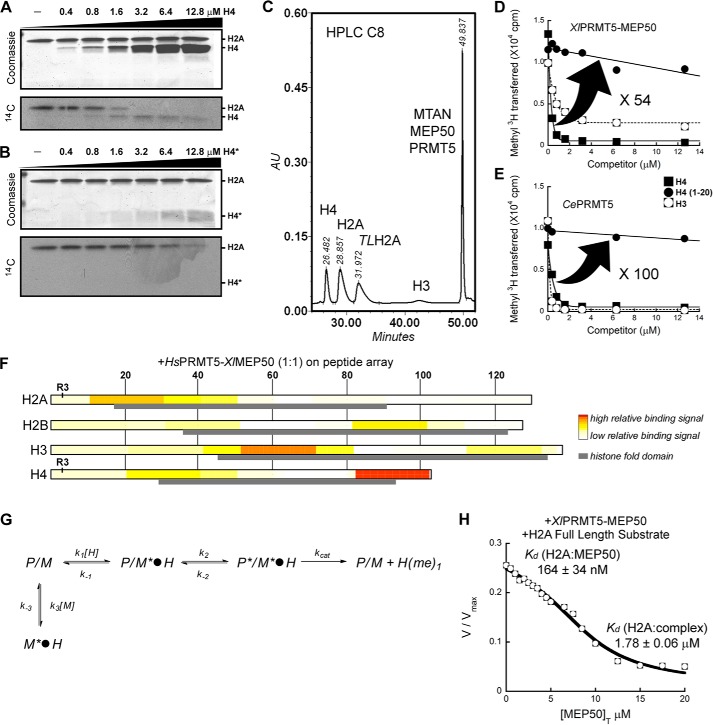
FIGURE 3.
XlMEP50 is a presenter that primarily binds histone H4 through the histone fold and exposes its N-terminal tail for methylation by XlPRMT5. A, substrate competition experiment where XlPRMT5-MEP50 activity toward histone H2A was displaced as histone H4 was titrated into the reaction (experiment performed using 50 mm MOPS, pH 7.0, 100 nm XlPRMT5-MEP50, 20 microunits of SeMTAN, 25 μm [14C]-methyl-SAM, and a H2A concentration kept constant at 2 μm while H4 was added at 0, 0.4, 0.8, 1.6, 3.2, 6.4, and 12.8 μm final concentrations). B, similar competition experiment as in A where peptide H4(1–20) was used as competing substrate (experimental conditions identical to those in A). C, quantification of methyl transfer reactions using reversed-phase HPLC. Histones H4, H2A, tailless H2A (TLH2A), and H3 were separated from each other and from radiolabeled substrate SAM using a C8 reverse HPLC column (see “Experimental Procedures”). D, substrate competition experiment where XlPRMT5-MEP50 activity toward histone H2A was displaced as histone H4, peptide H4(1–20), and histone H3 were titrated into the reaction (experiment performed using 50 mm MOPS, pH 7.0, 100 nm XlPRMT5-MEP50, 20 microunits of SeMTAN, 25 μm [3H]methyl-SAM, and a H2A concentration kept constant at 2 μm while competitors were added at 0–12.6 μm final concentrations). Transfer of [3H]methyl was quantified by liquid scintillation counting after isolation of histones. E, competition experiment similar to that in D where XlPRMT5-MEP50 was replaced by its homologous enzyme from C. elegans (experimental conditions identical to those in D). F, FLAG-HsPRMT5-XlMEP50 complex was incubated on ultrahigh density histone peptide arrays. PRMT5-MEP50 binding on the histone peptide scan data was extracted, and relative binding levels were plotted as a heat map, with no to low signal as white to light yellow, and high relative binding was plotted in red. Histone amino acid sequence numbers are represented at the top of the plots. The histone fold domain is indicated as a gray box, and the substrate residue Arg-3 (R3) on both H2A and H4 is indicated. G, relief of methyltransferase activity through the addition of MEP50. In this model used to determine the affinities of histone H2A for MEP50, P/M*·H represents the PRMT5-MEP50·histone complex, where only the histone fold is bound to the MEP50 presenter. P*/M*·H represents the PRMT5-MEP50·histone complex, where the histone fold is bound to the MEP50 presenter and the histone tail is bound to the enzyme active site. M*·H represents the complex between histone and exogenous MEP50. Each step is characterized by a constant (i.e. k1, k−1, k2, k−2, kcat, k3, and k−3). G, transferase reaction catalyzed by XlPRMT5-MEP50 was followed continuously at pH 7.7 using the luciferase-based assay with histone H2A as substrate (H2A fixed at 2 μm; see “Experimental Procedures”); XlMEP50 was added to the reactions (0–20 μm), and resulting transferase activities were recorded. Exogenous XlMEP50 competes with XlPRMT5-MEP50 for H2A binding, and methyl transfer is inhibited with increasing concentrations of XlMEP50. A dissociation constant (Kd) of H2A for both exogenous XlMEP50 (K′d H2A:MEP50) and PRMT5-associated MEP50 (Kd H2A:complex) was determined.