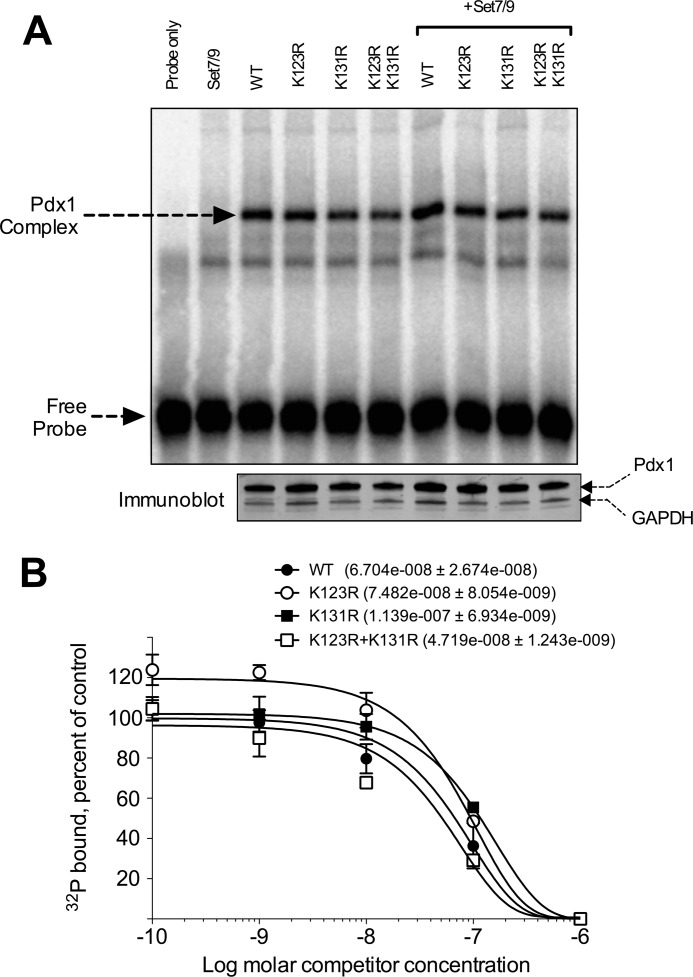
FIGURE 4.
Point mutations do not affect DNA binding by Pdx1. A, NIH3T3 cells were cotransfected with the WT or mutant Pdx1 proteins with or without cotransfection of Set7/9. Nuclear extracts from transfected cells were harvested and subjected to EMSA using the 32P-labeled E-A DNA fragment containing a Pdx1 binding site derived from Ins E2/A3/A4. The positions of the Pdx1-containing complex and the unbound, free probe are indicated by arrows. Bottom panel, immunoblots for Pdx1 and GAPDH from the same nuclear extracts as used for the EMSA. B, quantitation and modeling of EMSA reactions (as in A), with increasing concentrations of unlabeled E-A probe. For each Pdx1 mutant, the control condition is the fraction of total probe bound in the absence of competitor. The ordinate represents probe binding (at the given competitor concentration) as a percentage of the control condition. The apparent dissociation constants of the WT and mutant proteins are shown in the inset, with no statistical differences seen. The data shown are from three EMSA experiments.