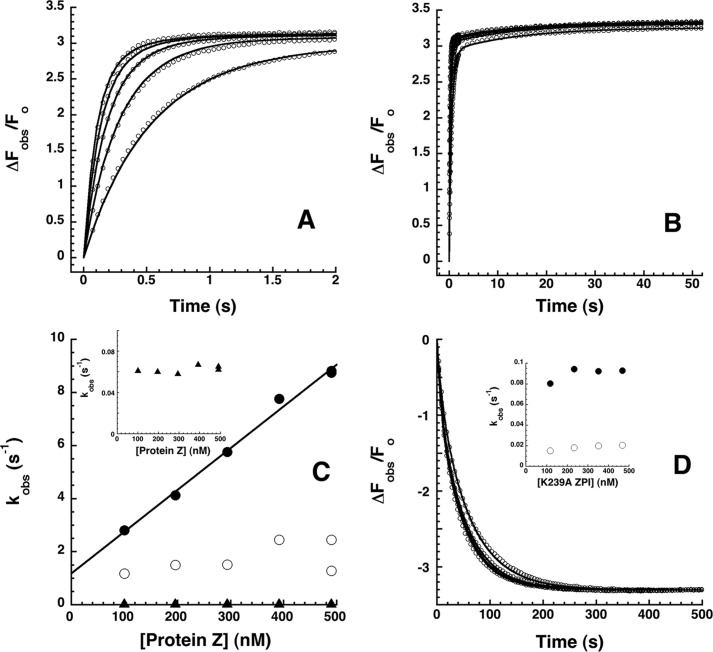
FIGURE 8.
Rapid kinetics of NBD-ZPI binding to PZ. A and B, NBD-ZPI (20 nm) was mixed with 100, 200, 300, 400, and 500 nm PZ, and the observed fluorescence changes (circles) were monitored over split time frames of 2 (A) and 50 s (B). Observed fluorescence changes from multiple averaged reaction traces were calculated by subtracting buffer background and fit by the three-exponential equation under “Experimental Procedures.” After correcting fluorescence amplitudes based on measured equilibrium KD values, ΔFobs was expressed relative to the fitted initial fluorescence. Every 10th data point is shown for clarity. Solid lines are global Kintek fits of complex association data together with dissociation data in D by a three-step sequential binding model. C, PZ concentration dependence of observed pseudo-first order rate constants for phase 1 (●), phase 2 (○), and phase 3 (▴) obtained from fits of data by the three-exponential function under “Experimental Procedures.” The solid line is a linear least squares fit of the phase 1 data, and the inset shows an expanded scale of the phase 3 data. D, kinetics of dissociation of the NBD-ZPI-PZ complex formed with 25 nm NBD-ZPI and 25 nm PZ after mixing with 200, 300, and 400 nm unlabeled K239A ZPI. Observed fluorescence changes were calculated as in A and B and fitted by the two-exponential equation under “Experimental Procedures.” After normalization based on the average fitted maximal fluorescence change, ΔFobs was expressed relative to the initial fluorescence of the complex. Solid lines are global Kintek fits of dissociation data together with the association data of A and B by the three-step binding model. The inset shows the K239A ZPI concentration dependence of kobs for phase 1 (●) and phase 2 (○).