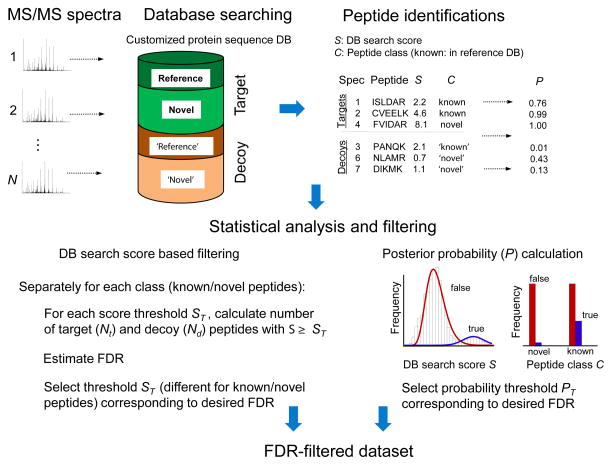
Figure 4. Statistical assessment of peptide identifications in proteogenomics.
MS/MS spectra are searched against a customized protein sequence database that includes target sequences for the organism of interest, i.e. a reference protein database and predicted protein sequences (containing novel peptides). In addition, two decoy databases (e.g. reversed sequences) of the same sizes as the target reference and predicted databases are appended to the target databases. The best database peptide match for each spectrum is selected for further analysis. Peptide identifications are classified as known or novel (for a decoy peptide the class - ‘known’ or ‘novel’ – is determined based on the class of the corresponding target sequence from which the decoy was generated). When using simple database search score based filtering, the numbers of target and decoy peptide identifications passing a certain score threshold are counted and used to estimate FDR corresponding to that threshold. FDR analysis should be done separately for known and novel peptides (class-specific FDR) due to difference in the number of known and novel sequences in the searched customized sequence database, and due to lower likelihood of correctly identifying a novel peptide than known peptide. When using more advanced methods based on computing posterior peptide probabilities, both the database search scores and the peptide class (known or novel) should be taken into consideration.