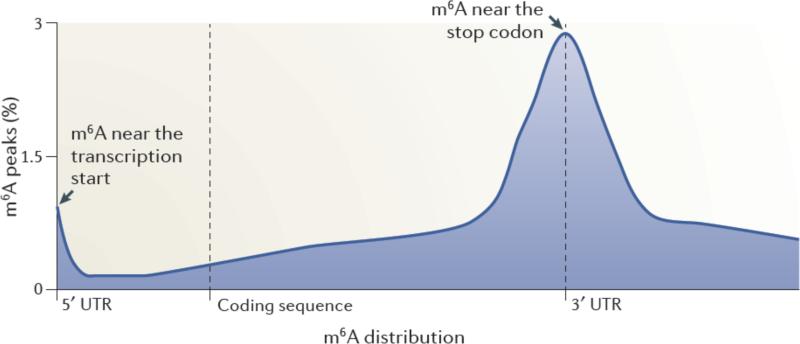
Figure 1. Spatially distinct pools of m6A in mRNAs.
Metagene analysis of MeRIP-Seq data shows that m6A is enriched in discrete regions of mRNA transcripts3. In this approach, each m6A peak in the transcriptome was mapped to a virtual transcript based on its position in the mRNA in which it is found. The metagene profile represents an overall frequency distribution of m6A residues along the entire mature transcript body. Shown is an idealized metagene profile based on published MeRIP-Seq datasets3. m6A residues can be found at the first encoded residue ,often as a dimethylated nucleotide (m6Am). m6A is also found at each of the other indicated positions in mRNAs, with a particularly prominent enrichment near the stop codon in mRNAs.