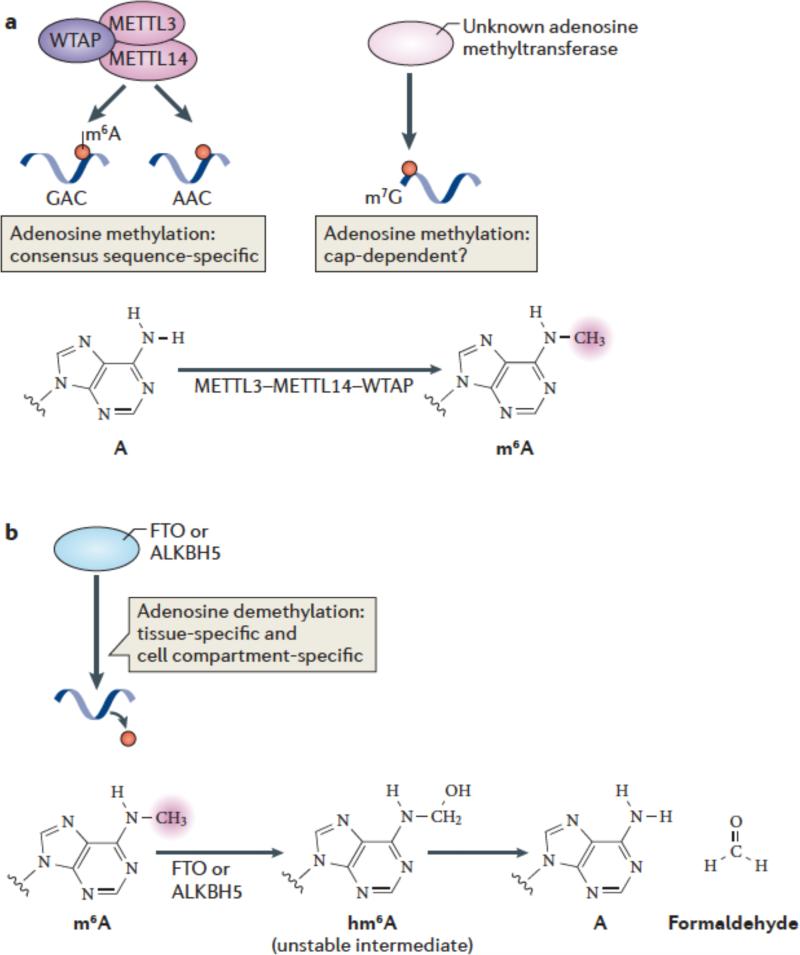
Figure 2. m6A methylation and demethylation pathways.
(a) METTL3 and METTL14 are two homologous m6A methyltransferases that synergize to methylate adenosines in RNA. WTAP is an additional component of this complex which lacks methyltransferase activity but which has a strong influence on m6A formation by interacting with METTL3/14. The central adenosines in the GAC and AAC motifs are the methylation sites of these enzymes. Additional methyltransferases may also contribute to m6A formation in cells, such as those which direct N6 methylation of adenosines associated with the cap structure. The generation of N6-methyladenosine (m6A) is shown.
(b) FTO and ALKBH5, the two mammalian m6A mRNA demethylases identified to date, catalyze methyl group removal from the N6 position of adenosine residues. FTO catalyzes oxidation of m6A in a reaction that requires O2, ascorbate, Fe(II) and 2-oxoglutarate. Oxidation of m6A generates CO2, succinate, and Fe(III). The product of this reaction is N6-hydroxymethyladenosine, an unstable intermediate that spontaneously decomposes to adenosine and formaldehyde. The squiggly lines refer to the position of the ribose sugar to which the base is attached.