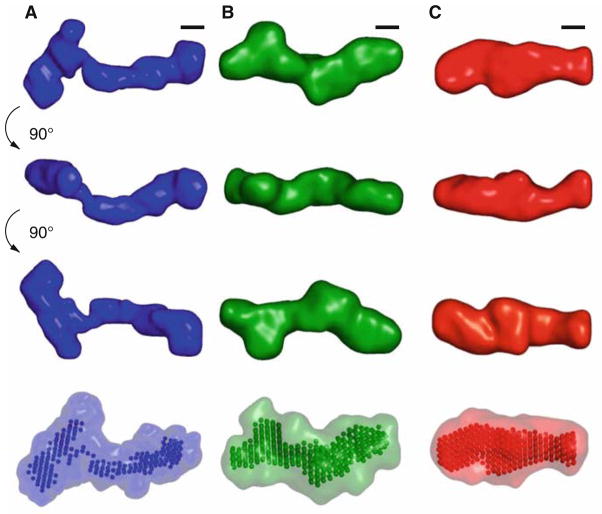
Fig. 3.
Low-resolution structural models for the VCI-II riboswitch tandem aptamer under different solution conditions. Average unfolded conformation (column A), conformation in the presence of 10 mM magnesium and absence of glycine (column B), and glycine-bound structure (column C). The first three rows show the “filtered” structure (see Subheading 3.6 for details of the reconstruction procedure) for each of the conformations in three different orientations. Black scale bars in each column correspond to 20 Å, the diameter of an A-form RNA helix. The rendered densities were generated by convoluting the bead models with a Gaussian kernel using the program Situs (46, 47). The last row shows the “filtered” models as beads and the convex hull of all bead models for a given conformation as a transparent surface. Reproduced from ref. 24 with permission from Elsevier Limited.