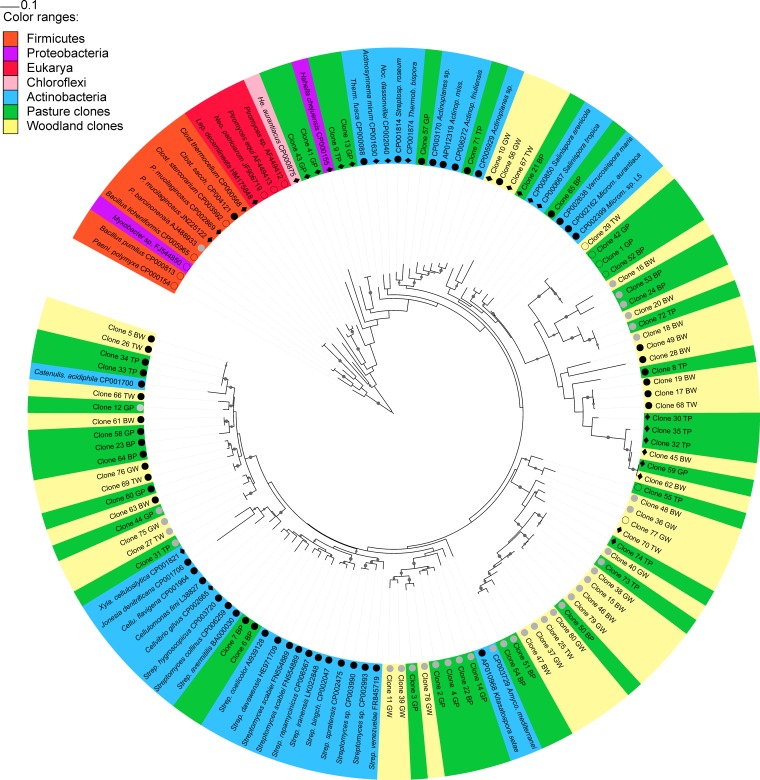
FIG 1.
Maximum likelihood tree (RaxML) constructed with GH48 sequences from soil clones and cultured strains from Actinobacteria, Firmicutes, Neocallimastigales (anaerobic fungi), Proteobacteria, Chloroflexi, and Insecta. Nodes in tree branches indicate bootstrap support of >0.8. Sequences from Bacillus spp. and Paenibacillus spp. were used as outgroups. GenBank accession numbers are indicated after the organism name. Colors indicate sequence taxonomy or soil clone provenance; symbols in front of sequence names indicate the presence or absence of aromatic amino acids relevant to cellulolytic action in T. fusca. Filled black circles indicate the presence of both amino acids, filled gray circles indicate the lack of phenylalanine 195, diamonds indicate the lack of tyrosine 213, and open circles indicate the lack of both aromatic amino acids. TW, Talmo woodland; GW, Glenrock woodland; BW, Bogo woodland; TP, Talmo pasture; GP, Glenrock pasture; BP, Bogo pasture; Clost. sacch, Clostridium saccharoperbutylacetonicum N1-4(HMT); Amyco, Amycolatopsis mediterranei S699; Actinop. miss, Actinoplanes missouriensis 431; Strep. bingch., Streptomyces bingchenggensis BCW-1.