Fig. 1.
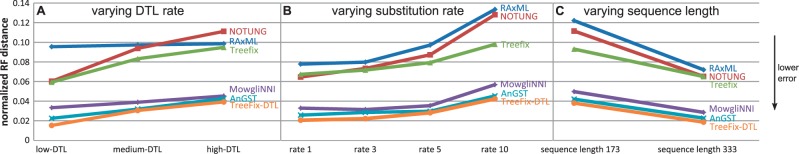
Error rates for different methods on simulated datasets. Error rates in terms of NRFD are shown for gene trees inferred using RAxML, NOTUNG, TreeFix, MowgliNNI, AnGST and TreeFix-DTL on the simulated 50-taxon datasets. Note that the lines are for visual guidance only. TreeFix-DTL produces the most accurate gene trees among all tested programs. (A) Results are shown for varying rates of DTL, averaged over all mutation rates and sequence lengths. (B) Results for varying mutation rates, averaged over all DTL rates and sequence lengths. (C) Results for varying sequence lengths, averaged over all DTL rates and mutation rates. Results for all 24 simulated datasets appear in Supplementary Figure S1
