Abstract
Motivation: Finding one or more cell populations of interest, such as those correlating to a specific disease, is critical when analysing flow cytometry data. However, labelling of cell populations is not well defined, making it difficult to integrate the output of algorithms to external knowledge sources.
Results: We developed flowCL, a software package that performs semantic labelling of cell populations based on their surface markers and applied it to labelling of the Federation of Clinical Immunology Societies Human Immunology Project Consortium lyoplate populations as a use case.
Conclusion: By providing automated labelling of cell populations based on their immunophenotype, flowCL allows for unambiguous and reproducible identification of standardized cell types.
Availability and implementation: Code, R script and documentation are available under the Artistic 2.0 license through Bioconductor (http://www.bioconductor.org/packages/devel/bioc/html/flowCL.html).
Contact: rbrinkman@bccrc.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Both flow cytometry (FCM) and mass cytometry are used to identify one or more cell populations of interest, such as those correlating with a disease phenotype. Cell types can be grouped by their particular combinations of markers, or immunophenotype. Despite progress made towards standardization of FCM assays (Maecker et al., 2012), the lack of a standard labelling strategy hampers data integration when trying to match gating data resulting from various manual or automated tools and heterogeneous sources. The cell ontology (CL) was developed to represent cell types relevant to biology. A recent revision (Diehl et al., 2011) specifically focused on extending coverage of the CL by improving the representation of haematopoeitic cell types, asserting those cell types in a single-inheritance hierarchy and adding logical restrictions to describe their immunophenotypes. However, multiple issues arose when we attempted to use the CL for labelling of cell types derived from FCM data. The definition of ‘mature T cell’ (obo:CL_0002419; “obo:” is an abbreviation for http://purl.obolibrary.org/obo/) mentions that it has plasma membrane part some ‘T cell receptor complex’ (obo:GO_0042101), which itself has part the CD3 marker (obo:PR_000001018), requiring complex reasoning to infer that ‘mature T cell’ has part some CD3. Additionally, when trying to identify CD3+ cells, FCM experimenters are in fact targeting CD3-epsilon (obo:PR_000001020), which is itself a subunit of the CD3 molecule described in the CL. As a consequence, while the CL encompasses information about surface markers, its manual exploitation can be challenging and may require multiple inferences along multiple axes, which is neither optimal nor sustainable with the increase in size of datasets. To address these challenges, we developed flowCL, a software package that enables researchers to unambiguously label their cell populations based on their immunophenotype using the CL. We also improved CL to enhance its utility for automated labelling of cell populations.
2 Approach
The core module of flowCL is an ontology labeller that attempts to provide a semantic identifier to a cell population based on its marker expression profile (i.e., the immunophenotype). The immunophenotype can be identified using either manual or automated methods (Malek et al., 2014; Finak et al., 2014; O’Neill et al., 2014; Qian et al., 2010). flowCL decomposes a query such as ‘CD4+CD8−’ (as provided in the conventional format used by immunologists) into its individual markers (CD4, CD8) and translates their relative abundance into a relation used in the CL, such as + for has plasma membrane part. The steps flowCL takes are as follows: (1) A SPARQL (http://www.w3.org/TR/rdf-sparql-query/) query against the CL fetches the labels and IDs corresponding to the input markers by text matching to the label or synonyms fields in the CL; (2) The marker labels are used to retrieve a list of cell types that contain (or lack) the marker labels; (3) The set of markers that make up each cell type is then retrieved; (4) A final query retrieves all parents up to the root of the CL for each cell type to build a tree diagram of the results.
3 Results and discussion
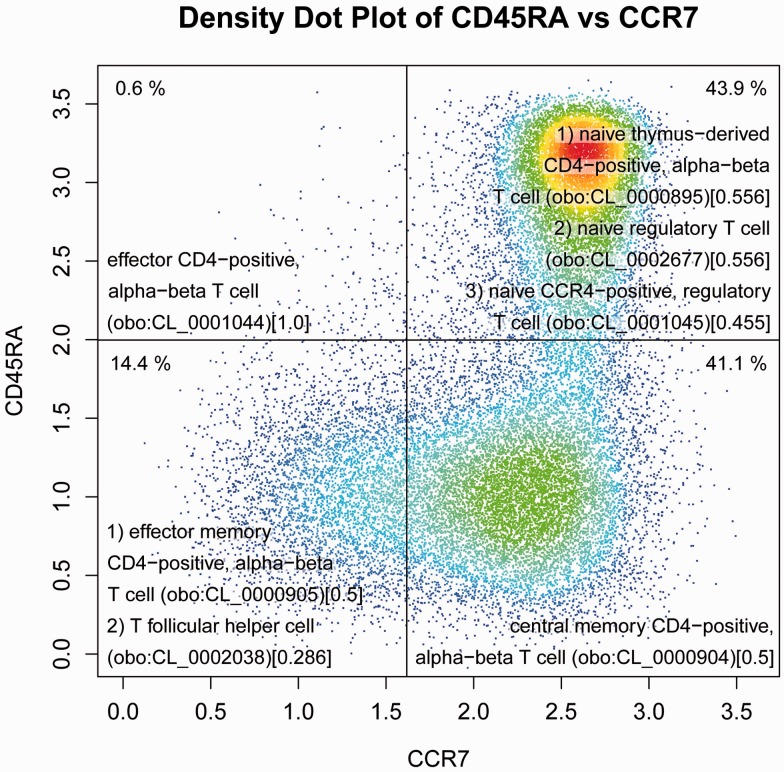
flowCL was successfully applied to label the cell populations from the Human Immunology Project Consortium (HIPC; Maecker et al., 2012). Figure 1 shows a density dot plot of CD45RA versus CCR7, created by a combination of manual analysis and flowDensity (Malek et al., 2014). Using flowCL and the immunophenotype ‘CD3+CD4+CD8-CCR7-CD45RA+’, an exact match (all markers of the input immunophenotype, along with their specific abundance, correspond to all of the markers that make up the cell type’s definition in CL) was returned and the population was labelled ‘effector CD4-positive, alpha-beta T cell’ (Fig. 1, top left quadrant). The top right quadrant of Figure 1 shows where three possible labels are returned when the same specific combination of markers are queried. In this case, flowCL returns all possible matches and computes a ranking score to indicate relevance to the original query. More detail on the flowCL algorithm and results is available in the Supplementary Material.
Fig. 1.
Density dot plot of CD45RA versus CCR7 created using flowDensity and using data from Cambridge Institute for Medical Research, after pre-gating to identify live cells, lymphocytes, CD3+, CD4+ and CD8−. Each quadrant has the cell labels that were returned by flowCL for each respective cell population, followed by the CL identifier in parentheses and ranking score in square brackets
During the development of flowCL, we made several changes to the CL to add synonyms of markers, as well as new restrictions on existing cell types to account for the way they are identified. The current version of flowCL relies on the CL as the ontology source as this is, to the best of our knowledge, the only resource describing cell markers logically for haematopoeitic cell lines. Our code could however be adapted to use another resource; the SPARQL queries are stored in a separate function file from the main code, and only require the URL of a SPARQL endpoint (http://cell.ctde.net:8080/openrdf-sesame/repositories/CL is used for flowCL) as a parameter.
4 Conclusion
Decoupling the knowledge representation and the software code allows for greater flexibility for extension and updates. As the CL development proceeds, flowCL automatically remains up-to-date with the latest scientific knowledge. Additionally, the information added to the CL is immediately available for the scientific community, independently of the flowCL development or the HIPC project. By providing automated labelling of cell populations based on their immunophenotype, flowCL allows researchers to report results using cell type labels. Future work includes validation on other datasets and extending the CL with an assay-based representation of cell types.
Funding
This work was supported by National Institutes of Health (NIH)/National Institute of Biomedical Imaging and Bioengineering (NIBIB) [R01 EB008400], Human Immunology Project Consortium (HIPC) [U19 AI089986], Natural Sciences and Engineering Research Council of Canada, National Institute of General Medical Sciences (NIGMS) 2R01GM080646-06 (Protein Ontology) and HHSN272201200028C (ImmPort).
Conflict of Interest: None declared.
Supplementary Material
References
- Diehl A.D., et al. (2011). Hematopoietic cell types: prototype for a revised cell ontology. J. Biomed. Inf., 44, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., et al. (2014). OpenCyto: an open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol., 10, e1003806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maecker H.T., McCoy J.P., Nussenblatt R. (2012). Standardizing immunophenotyping for the human immunology project. Nat. Rev. Immunol., 12, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek M., et al. (2014). flowDensity: reproducing manual gating of flow cytometry data by automated density-based cell population identification. Bioinformatics: btu677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill K., et al. (2014). Enhanced flowType/Rchyoptimyx: a bioconductor pipeline for discovery in high-dimensional cytometry data. Bioinformatics, 30, 1329–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., et al. (2010). Elucidation of seventeen human peripheral blood b-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry Part B, 78, S69–S82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.