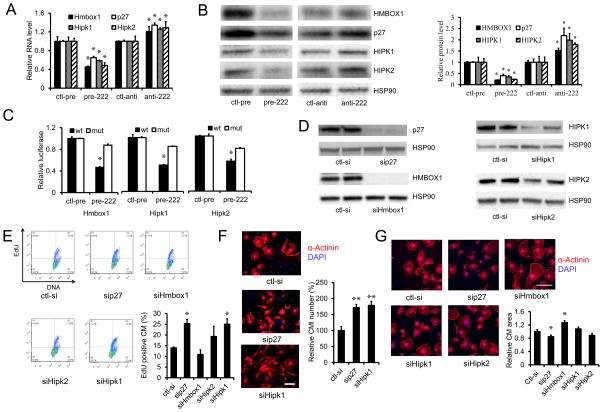
Figure 3. miR-222 targets in cardiomyocytes.
A and B. qRT-PCR and immunoblotting were used to analyze RNA and protein levels of the four putative miR-222 targets in neonate cardiomyocytes treated with control precursor (ctl-pre), miR-222 precursor (pre-222), control antimiR (ctl-anti), or antimiR-222 (anti-222), respectively. Data are shown as fold-change in gene expression normalized to U6 in (A). These data demonstrate that miR-222 decreases RNA and protein levels for all four targets in primary cardiomyocytes. C. Luciferase assays of COS7 cells co-transfected with control precursor (ctl-pre) or miR-222 precursor (pre-222) and reporter plasmids containing 3′UTR wild-type or mutated miR-222 binding sites for each of the putative target genes. These data demonstrate all three candidates are direct targets of miR-222. D. Immunoblotting of the four genes in neonate cardiomyocytes transfected with control siRNA (ctl-si) or siRNAs of p27 (sip27), Hmbox1 (siHmbox1), Hipk2 (siHipk2) and Hipk1 (siHipk1) demonstrate effective knock-down for all. HSP90 was used as a loading control. E. Flow cytometry for EdU in neonate cardiomyocytes transfected with indicated siRNAs demonstrates that knockdown of p27 or HIPK1 increases EdU incorporation in cultured cardiomyocytes, consistent with proliferation. F and G. Neonate cardiomyocytes cultures were stained for sarcomeric α-actinin to identify cardiomyocytes, and cardiomyocyte number and area were quantified. Knockdown of p27 and HIPK1 increases cardiomyocyte proliferation (F) while Hmbox1 knockdown increases cardiomyocytes size (G). At least 200 cells or 30 images were quantified in each group. Data represent the mean±SEM from at least three independent experiments. Scale bar: 100 μm. *p<0.05, **p<0.01 versus respective control using Student’s test.