Abstract
Genetic fusion of two proteins frequently induces beneficial effects to the proteins, such as increased solubility, besides the combination of two protein functions. Here, we study the effects of the bacterial surface layer protein SgsE from Geobacillus stearothermophilus NRS 2004/3a on the folding of a C-terminally fused enhanced green fluorescent protein (EGFP) moiety. Although GFPs are generally unable to adopt a functional confirmation in the bacterial periplasm of Escherichia coli cells, we observed periplasmic fluorescence from a chimera of a 150-amino-acid N-terminal truncation of SgsE and EGFP. Based on this finding, unfolding and refolding kinetics of different S-layer-EGFP chimeras, a maltose binding protein-EGFP chimera, and sole EGFP were monitored using green fluorescence as indicator for the folded protein state. Calculated apparent rate constants for unfolding and refolding indicated different folding pathways for EGFP depending on the fusion partner used, and a clearly stabilizing effect was observed for the SgsE_C fusion moiety. Thermal stability, as determined by differential scanning calorimetry, and unfolding equilibria were found to be independent of the fused partner. We conclude that the stabilizing effect SgsE_C exerts on EGFP is due to a reduction of degrees of freedom for folding of EGFP in the fused state.
Keywords: S-Layer protein, GFP, fusion protein, protein folding, periplasm
Genetically fused moieties in a fusion protein can exhibit mutual effects on the function of either part. Much effort has been undertaken to identify fusion tags that might increase the yield of correctly folded recombinant proteins by enhancing the expression and solubility, or preventing proteolytic degradation [1]. However, any identified effects are dependent on both the fusion tag and the fused target protein [7], and the influence on proper folding is exerted by keeping the protein in solution [18]. Folding might be improved directly by co-expression of chaperones [13]. Another fusion protein approach that intervenes with protein folding itself has been proposed through N-terminal fusion of target proteins to RNA-binding protein, which in turn attracts RNA that exerts chaperoning function on the fused protein [2].
Here, we report on directly improved folding properties of the model protein enhanced green fluorescent protein (EGFP) [23, 27], depending on the fusion to a truncated surface (S)-layer protein SgsE from the Gram-positive bacterium Geobacillus stearothermophilus NRS 2004/3a.
The intrinsic function of S-layer proteins is to self-assemble into a 2D crystalline layer surrounding the entire bacterial cell, providing stability to the cell as well as an additional barrier to the environment [24]. Technical applications of S-layer protein assemblies as a matrix for fused or chemically conjugated biologically active molecules (e.g., enzymes, antigenic epitopes, carbohydrates) have been investigated [14, 30]. Insight into the effects of S-layers on closely attached proteins might increase their applicability as well as strengthen our general knowledge about protein environments.
The enhanced GFP variant used in this study contains the two well-studied chromophore mutations: S65T, leading to brighter fluorescence with a single excitation peak at 490 nm; and F64L, allowing stable folding at 37°C [3, 11]. Folding of GFP was shown to be affected by fused proteins that are prone to misfolding and aggregation, enabling the use of GFP as a “folding reporter” [28]. Green fluorescence is dependent on the intact tertiary structure that shields and stabilizes the chromophore, making the fluorescence signal a direct indicator of the folded state of GFP [9].
It was shown that GFP is inactive in the periplasmic space of E. coli cells when exported in fusion to maltose-binding protein (MBP) following a general secretory (Sec-dependent) export pathway for unfolded proteins [8]. Lack of periplasmic fluorescence was attributed to the inability of GFP to fold correctly in the periplasm. Later on, this hypothesis was supported by the work of Thomas et al. [26] showing that GFP could be exported in a folded and active state to the periplasm in E. coli following the twin-arginine translocation (TAT) pathway. The general inability of GFP to function in the periplasm is frequently used as a reporter feature for the orientation of membrane proteins [4].
Here we report on the fusion partner SgsE_C, which is a 150-amino-acid N-terminal truncation of SgsE, that increases the unfolding and refolding stability of GFP in vitro and allows, for the first time, for detection of fluorescence from a green fluorescent protein in a chimeric state that was exported to the periplasm of E. coli host cells via a Sec-dependent pathway.
Materials and Methods
Construction of Genetic Fusion Vectors
Vectors allowing for cytoplasmic and periplasmic expression of the proteins of interest were constructed by standard molecular methods and using E. coli DH5a and heat-shock transformation. All PCR (polymerase chain reaction) primer sequences used are listed in Table 1.
Table 1.
PCR primers used in this studya.
| Primer | Nucleotide sequence (5′ → 3′) |
|---|---|
| EGFP_XhoI_rev | AATCACTCGAGTTACTTGTACAGCTCGTCCATGC |
| EGFP-rev-HindIII | GATGAAGCTTTCACTTGTACAGCTCGTCCATG |
| EGFP-BamHI/NcoI for | TGTAGGATCCATGGTGAGCAAGGGCG |
| SgsE_G_NcoI_for | AACTGCCATGGTTACGCTCAAAGCCATTGATG |
| SgsE_M_NcoI_for | GTATCCATGGTACTTAAGGCTGTCGGCGC |
| malE_EGFP-overlap-for | GCGCAGACTCGTATCACCAAGGTACCGGTGAGCAAGGGCGAGGAGC |
| malE_EGFP-overlap-rev | GCTCCTCGCCCTTGCTCACCGGTACCTTGGTGATACGAGTCTGCGC |
| NdeI-malE_with_SP_for | GAGAACATATGAAAATAAAAACAGGTGC |
| NdeI-malE_wo_SP_for | GTCAACATATGGAAGAAGGTAAACTGGTAATCTGG |
Restriction sites are underlined; ATG start codons are written in bold letters.
The SgsE forms used were SgsE_C (residues 133–903), ranging from amino acid residue 133 to the native C-terminus at residue 903, SgsE_G (residues 331–903), and SgsE_M (residues 609–903). Cloning of sgsE_C, egfp, and sgsEC-egfp into the expression vector pET28a, yielding pET28a-SgsE_C, pET28a-EGFP, and pET28a-SgsE_C-EGFP, respectively, was described previously [15, 21].
sgsE_G-egfp and sgsE_M-egfp were PCR-amplified from sgsEC-egfp, using the primer pairs SgsE_G_NcoI_for / EGFP_XhoI_rev and SgsE_M_NcoI_for / EGFP_XhoI_rev, respectively. The PCR products were cloned into pET28a via NcoI and XhoI, yielding pET28a-SgsE_G-EGFP and pET28a-SgsE_M-EGFP.
egfp, sgsEC-egfp, sgsEG-egfp, and sgsEM-egfp were subcloned from the respective pET28a constructs into pET22b via NcoI and XhoI immediately downstream of the pelB signal sequence for periplasmic targeting, yielding pET22b-(PelB)EGFP, pET22b-(PelB)SgsE_C-EGFP, pET22b-(PelB)SgsE_G-EGFP, and pET22b-(PelB)SgsE_M-EGFP.
A fusion sequence of the MBP gene malE and egfp was obtained by overlap extension PCR, including the native MBP-signal peptide. The primer pairs NdeI-malE_with_SP_for / malE_EGFP-overlap-rev and malE_EGFP-overlap-for / EGFP-rev-HindIII were used to amplify malE and egfp. A PCR mixture containing these two products and the primers NdeI-malE_with_SP_for and EGFP-rev-HindIII was used to amplify the final fusion product. A fusion sequence lacking the signal peptide and starting at codon 29 of malE-egfp was obtained similarly, using the primer NdeI-malE_wo_SP_for instead of NdeI-malE_with_SP_for. Both sequences were cloned into pET22b via NdeI and HindIII restriction sites, thus replacing the pelB leader sequence originally contained in pET22b, yielding pET-(SP)MBP-EGFP and pET-MBP-EGFP.
Growth of Transformed E. coli Cells
E. coli BL21 cells were transformed with the constructs described above. Transformed cells were grown in Luria–Bertani medium at 37°C to the mid-exponential growth phase, and protein expression was induced with a final concentration of 0.5 mM isopropyl-β-d-thiogalactopyranoside. Expression cultures used for protein purification were subsequently grown for 4 h at 30°C. Cultures used for fluorescence microscopy and transmission electron microscopy were grown at 37°C for 3 h. All cultures were incubated with shaking at 200 rpm. Ampicillin and kanamycin were added to the medium at a concentration of 100 μg/ml and 50 μg/ml, respectively, when appropriate.
Isolation and Purification of Chimeras
EGFP and SgsE-EGFP fusion proteins were purified as described previously using size-exclusion chromatography [21]. MBP-EGFP was bound to an amylose column (XK16 column 1.6 × 10 cm; GE Healthcare Life Sciences) in buffer A (20 mM Tris/HCl, pH 7.5, containing 0.2 M NaCl, 1 mM EDTA, and 5 mM β-mercaptoethanol) and eluted using buffer A containing 10 mM maltose. SgsE_G-EGFP and SgsE_M-EGFP were further purified by Uno Q (Bio-Rad) anion-exchange chromatography in 50 mM Bis-Tris, pH 6.5, and elution with increasing NaCl concentration ranging from 50 mM to 1 M. Fluorescent fractions were collected and dialyzed against an excess of sample buffer (50 mM Tris/HCl, pH 8.0, 2 mM EDTA), at 4°C.
The final protein concentrations of the purified samples were calculated from the absorbance at 280 nm and the calculated molar extinction coefficient derived from the amino acid sequence of the individual protein constructs, using ProtParam [10].
Western Immunoblotting
SDS-PAGE and subsequent Western blotting were performed as described previously [30], using mouse anti-GFP (Roche) as primary antibody and IRDye 800CW Goat Anti-Mouse antibody (LI-COR) as secondary antibody. Detection was performed at 800 nm using the Odyssey imaging system (LI-COR).
Fluorescence Microscopy
Ten μl of E. coli BL21 cell suspensions expressing the various constructs was mixed with 10 μl of solution of molten 0.9% agarose, 40 mM Tris, 20 mM acetic acid, 1 mM EDTA at pH 8.0, and pre-heated to 55°C, followed by immediate covering of the sample with a cover glass to immobilize the cells in the solidified agarose. The samples were then monitored using a Nikon Eclipse TE2000-S fluorescence microscope at a magnification of 100 × using an oil immersion objective and a GFP filter. Pictures were taken with a Nikon Digital Sight DS-Qi1Mc camera and the NIS-Elements 3.22 imaging software.
Immunogold Labeling and Transmission Electron Microscopy
E. coli cells expressing PelB-SgsE_C-EGFP were fixed in cacodylate buffer containing p-formaldehyde and glutaraldehyde and embedded in LR-white resin (Sigma-Aldrich) as described previously [25]. The resin block was trimmed on a Leica EM TRIM2 and cut on a Leica EM UC7 to thin sections of approximately 50 nm thickness.
The thin sections were immunogold-labeled using rabbit anti-SgsE antiserum as primary antibody and goat anti-rabbit antibody conjugated to 5 nm gold particles (Sigma-Aldrich) as described previously [25]. Electron micrographs of the immunogold-labeled, thin-sectioned cells were taken with a FEI Tecnai G2 20 transmission electron microscope at 80 keV with a FEI Eagle 4k camera at 50,000 × magnification.
Protein Unfolding and Refolding
Unfolding and refolding measurements were performed on the chimeras SgsE_C-EGFP, SgsE_G-EGFP, SgsE_M-EGFP, and MBP-EGFP, and on EGFP at 22°C. Green fluorescence served as a probe for correct folding of EGFP [9] and was measured in a Perkin Elmer Instruments LS55 Luminescence Spectrometer at an emission wavelength of 509 nm and excitation at 488 nm. The bandwidth for both excitation and emission was 5 nm. All unfolding and refolding measurements were made at a final protein concentration of 0.05 μM in sample buffer containing guanidinium hydrochloride (GdmCl) at the defined concentrations described below. Fluorescence was measured every 0.01 min and samples were stirred at low stirring speed to ensure homogeneity under continuous local excitation.
To monitor unfolding kinetics, the protein solution was diluted 1:200 with sample buffer containing 3 M GdmCl, and fluorescence was monitored for 90 to 100 min.
For refolding measurements, protein samples were diluted 1:20 in sample buffer containing 6 M GdmCl, resulting in a final GdmCl concentration of 5.7 M. Samples were incubated for 2 h under rotation at 22°C, leading to a solution of unfolded protein. This solution was diluted 1:10 in sample buffer, mixed thoroughly, and subjected immediately to fluorescence measurement. Fluorescence was monitored for 60 min. All measurements were repeated three times.
Unfolding and Refolding Data Analysis
The obtained unfolding and refolding curves were fitted to appropriate exponential models using the Gauss–Newton algorithm implemented in the statistics software R 2.10.
Unfolding curves were fitted to the bi-exponential decay function
where y is the fluorescence signal, t is the elapsed time, and ku1 and ku2 are the apparent rate constants for unfolding. The pre-exponential coefficients A0, A1, and A2 were normalized to give a sum of 1, and the original data were normalized with the same factor. Normalized data sets for each sample were then globally fitted to the above model.
Refolding data were normalized to the original fluorescence values of untreated samples. The resulting curves of each sample data set were globally fitted to the exponential equation
with kf1 and kf2 being the apparent rate constants for refolding, and ku being the apparent rate constant for differently pronounced unfolding taking place throughout the measurement. Nonsignificant parameters were removed in the final models.
Equilibrium Unfolding
Protein samples were incubated at a concentration of 0.05 μM for 24 h at 22°C in sample buffer containing increasing concentrations of GdmCl (0.00, 1.00, 2.00, 2.66, 2.83, 3.00, 3.16, 3.33, 3.66, 4.00, 4.33, 4.66, 5.00, and 6.00 M). After incubation, green fluorescence was excised at 488 nm and detected at 509 nm in a Perkin Elmer Instruments LS55 Luminescence Spectrometer. The bandwidth for excitation and emission was 5 nm. Fluorescence values were normalized to an average of three measurements of the respective sample at 0 M GdmCl and plotted against the corresponding values of GdmCl concentration. For each resulting curve, the transition midpoint was calculated as the inflection point from a fitted bi-sigmoidal model curve.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) was performed using an automated VP-Capillary DSC Microcalorimeter (MicroCal), using protein solutions of approximately 7 μM in sample buffer. The temperature interval was from 20°C to 110°C and the heating rate was 1°C/min. Buffer baselines (obtained from reheating of the denatured sample solutions) were subtracted, and data were normalized for protein concentration and fitted with a non-2-state thermal unfolding model using the software supplied by MicroCal (Origin 7).
Results
Periplasmic Activity of EGFP Fused to SgsE
The chimeras (PelB)SgsE_C-EGFP, (PelB)SgsE_G-EGFP, and (PelB)SgsE_M-EGFP as well as sole (PelB)EGFP were expressed with an N-terminal PelB signal peptide, and (SP)MBP-EGFP was expressed with its native signal peptide, mediating transfer to the periplasm in E. coli BL21 cells following a Sec-dependent pathway. Protein expression was verified by Western blotting using anti-EGFP antibody (Fig. S1), and the activity and location of EGFP were monitored by fluorescence microscopy. Periplasmically targeted (PelB)SgsE_C-EGFP was observed to be fluorescent, most profoundly at the edges of the E. coli BL21 host cells (Fig. 1A). The other chimeras and similarly targeted control proteins (SP)MBP-EGFP and (PelB)EGFP did not show any periplasmic fluorescence; the same proteins expressed without the periplasmic targeting signal gave strong cytoplasmic fluorescence signals in all cases (data not shown).
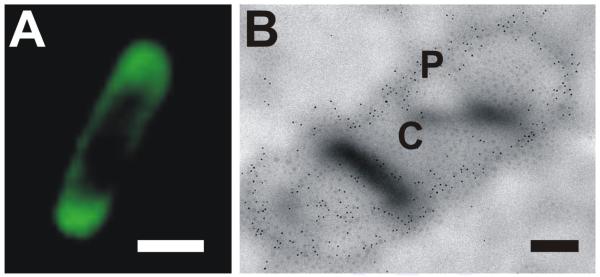
Fig. 1.
Fluorescence micrograph (A) and transmission electron micrograph after immunogold labeling and thin-sectioning (B) of E. coli BL21 cells expressing (PelB)SgsE_C-EGFP.
P, periplasm; C, cytoplasm. The SgsE moiety was detected with rabbit anti-SgsE antibody and goat anti-rabbit antibody conjugated to 5 nm gold particles. The fusion protein was found to reside almost exclusively in the periplasm. The scale bars represent 1 μm (A) and 200 nm (B).
To verify the periplasmic localization of (PelB)SgsE_C-EGFP, E. coli expression cells were embedded, thin-sectioned, and immunogold-labeled with rabbit anti-SgsE antibody and gold-conjugated goat anti-rabbit antibody. Immunogold-labeled thin-sectioned cells were examined by transmission electron microscopy, detecting SgsE moieties almost exclusively in the periplasmic space (Fig. 1B).
This observation led us to conclude that SgsE_C had a beneficial effect on the folding of its EGFP fusion partner, allowing it to adopt a functional conformation under otherwise detrimental conditions.
Protein Unfolding Kinetics
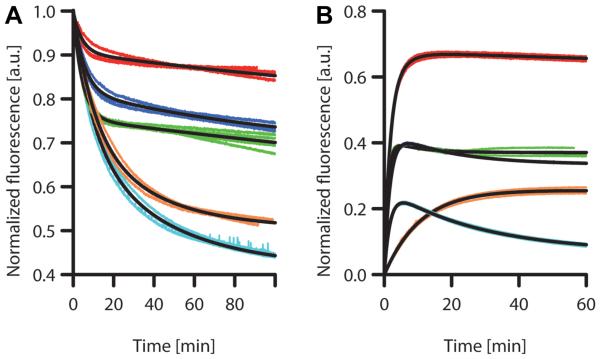
Based on the above findings, we investigated the impact of the fusion of EGFP to different SgsE forms and to MBP on its unfolding and folding kinetics. The intensity of green fluorescence at 509 nm served as a probe for the folded state of EGFP [9]. The emission and excitation spectra of fused and sole EGFP were shown to be identical [15]. Unfolding measurements were performed on EGFP and EGFP fused to the SgsE forms SgsE_C, SgsE_G, SgsE_M, as well as to MBP in 3 M GdmCl, which corresponds to a denaturant concentration at which a significant amount of all fusion proteins is in the unfolded state. Resulting biphasic unfolding time traces and fitted model curves are displayed in Fig. 2A, and the corresponding coefficients are listed in Table 2. The apparent rate constants are sorted with ku1 being greater than ku2. Thus, Au1·exp(−ku1·t) describes the fast and Au2·exp(−ku2·t) the slower decrease of the observed signal.
Fig. 2.
Protein unfolding in 3 M GdmCl (A) and refolding measurements after rapid dilution from 5.7 M to 0.57 M GdmCl (B).
Data curves are displayed for SgsE_C-EGFP (red), SgsE_G-EGFP (blue), EGFP (green), MBP-EGFP (orange), and SgsE_M-EGFP (cyan). Fitted curves are displayed in black color.
Table 2.
Coefficients ± standard errors for protein unfolding, assuming a bi-exponential process.
| Apparent rate constants (min−1)*100 | Pre-exponential coefficients*100 | ||||
|---|---|---|---|---|---|
|
| |||||
| Protein | ku1 | ku2 | A0 | Au1 | Au2 |
| SgsE_C-EGFP | 23.34 ± 0.17 | 0.44 ± 0.01 | 75.57 ± 0.38 | 9.15 ± 0.03 | 15.05 ± 0.36 |
| SgsE_G-EGFP | 18.99 ± 0.10 | 0.84 ± 0.02 | 67.39 ± 0.15 | 18.33 ± 0.05 | 14.47 ± 0.13 |
| SsgE_M-EGFP | 10.29 ± 0.08 | 2.35 ± 0.02 | 41.50 ± 0.05 | 28.50 ± 0.23 | 29.63 ± 0.20 |
| MBP-EGFP | 9.63 ± 0.08 | 2.75 ± 0.03 | 50.37 ± 0.03 | 26.96 ± 0.28 | 22.45 ± 0.27 |
| EGFP | 22.66 ± 0.10 | 0.22 ± 0.02 | 47.57 ± 2.68 | 24.63 ± 0.06 | 28.00 ± 2.65 |
Fusion to SgsE_C significantly decreased both the rate and extent of unfolding of EGFP, whereas further N-terminal truncation of the fusion partner protein (SgsE_G) displayed only a slightly increased stability compared with sole EGFP. By contrast, fusion to SgsE_M and MBP decreased the conformational stability of EGFP. Both SgsE-M-EGFP and MBP-EGFP showed different overall unfolding characteristics, specified by significantly lower values for ku1 and higher values for ku2 when compared with the other three chimeras under examination. The extent of unfolding calculated from the loss of fluorescence intensity for MBP-EGFP and SgsE_M-EGFP in the monitored time range of 100 min was 51% and 43%, respectively. This compares with 68% and 72% for EGFP and SgsE_G-EGFP, and 84% for SgsE_C-EGFP.
Protein Refolding Kinetics
Protein refolding measurements were performed in a separate experimental set-up by dilution of solutions of unfolded protein (final concentration of 0.05 μM) leading to a decrease from 5.7 M GdmCl to 0.57 M GdmCl. Green fluorescence served as a probe for the folded state of EGFP. Fig. 2B shows original time traces and fitted model curves. The corresponding apparent rate constants and pre-exponential coefficients are listed in Table 3. All three SgsE-EGFP chimeras exhibited comparable values for kf1, which describes the fast refolding process. In contrast, refolding of sole EGFP showed a higher kfl, reflecting faster refolding, whereas MBP-EGFP refolded slowly.
Table 3.
Coefficients ± standard errors for protein refolding, assuming bi- or tri-exponential processes.
| Apparent rate constants (min−1)*100 | Pre-exponential coefficients*100 | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Protein | kf1 | kf2 | ku | A0 | Af1 | Af2 | Au |
| SgsE_C-EGFP | 62.58 ± 0.61 | 25.30 ± 0.55 | 0.05 ± 0.00 | −50.17 ± 0.77 | −16.85 ± 0.80 | 67.60 ± 0.02 | |
| SgsE_G-EGFP | 58.56 ± 0.09 | 5.51 ± 0.02 | 33.41 ± 0.01 | −43.39 ± 0.03 | 10.38 ± 0.02 | ||
| SsgE_M-EGFP | 57.87 ± 0.19 | 3.57 ± 0.01 | 6.78 ± 0.03 | −25.99 ± 0.05 | 19.53 ± 0.02 | ||
| MBP-EGFP | 9.33 ± 0.03 | 0.03 ± 0.00 | −26.04 ± 0.04 | 25.96 ± 0.05 | |||
| EGFP | 96.60 ± 0.69 | 12.01 ± 0.32 | 37.06 ± 0.01 | −40.60 ± 0.15 | 4.35 ± 0.11 | ||
Folding curves were best fitted by an exponential equation containing two apparent rate constants for (re)folding (kf1 and kf2) and one for concomitant unfolding (ku). For the description of the refolding process of the SgsE_C-EGFP chimera both kf1 and kf2 were important, whereas in the case of the other chimeras, kf2 could be neglected. This indicates a more complex refolding pathway for SgsE_C-EGFP.
The apparent rate constant ku, which accounts for concomitant unfolding, was extremely small for SgsE_C-EGFP and MBP-EGFP (Table 3), indicating the formation of stably refolded protein forms. SgsE_G-EGFP and SgsE_M-EGFP showed smaller ku values than EGFP; however, the corresponding pre-exponential coefficient Au was more than a factor of 2 larger, pointing to an overall larger extent of concomitant unfolding in these two chimeras.
The extent of refolding is reflected by A0 and, in the case of low ku values, also by Au. SgsE_C-EGFP showed the largest extent of refolding of all tested constructs, peaking at 67% of the initial fluorescence value. SgsE_G-EGFP and EGFP exhibited a comparable fluorescence recovery of 40% and 39%, respectively. Upon refolding of SgsE_M-EGFP, only 22% of the initial fluorescence was regained, whereas MBP-EGFP reached 25% refolding, albeit only after 50 min.
The shape and timescale of the obtained unfolding and refolding curves as well as the apparent rate constants determined for refolding of EGFP compare well with the data reported in a comprehensive work by Xie and Zhou [29].
Protein Unfolding Equilibria
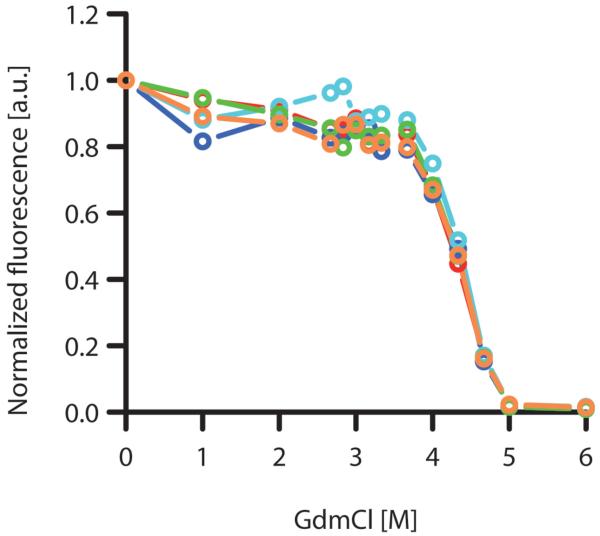
The extent of unfolding after 24 h of incubation in increasing concentrations of GdmCl was measured as the green fluorescence intensity relative to a base fluorescence value of samples, which had been incubated in the absence of denaturing GdmCl. The observed unfolding equilibria curves of the EGFP moiety in SgsE_C-EGFP, SgsE_G-EGFP, SgsE_M-EGFP, MBP-EGFP, and sole EGFP did not show significant differences (Fig. 3). The respective transition curve midpoints were found at 4.34, 4.35, 4.35, 4.36, and 4.36 M GdmCl.
Fig. 3.
Unfolding transition curves of SgsE_C-EGFP (red), SgsE_G-EGFP (blue), EGFP (green), MBP-EGFP (orange), and SgsE_M-EGFP (cyan).
Samples were incubated for 24 h in buffer containing the indicated GdmCl concentration. The data points are connected by straight lines.
Differential Scanning Calorimetry
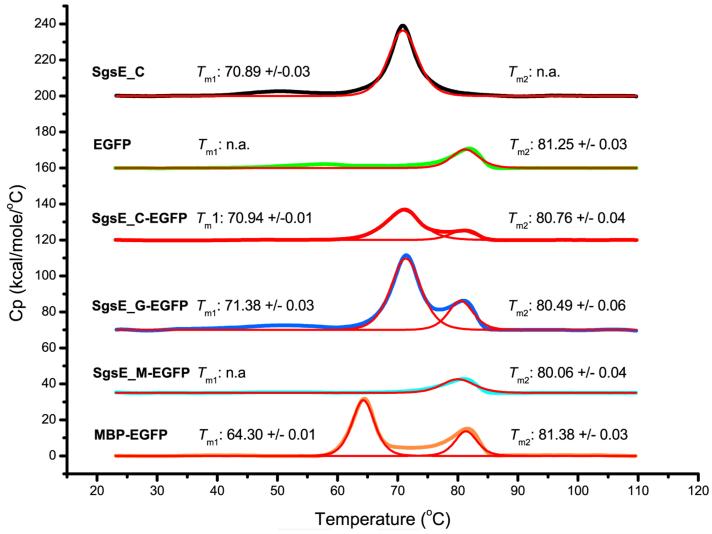
To identify possible effects of the translational fusion on the thermal stability of either fusion moiety, differential scanning calorimetry (DSC) was conducted on SgsE_C and EGFP as well as on the chimeras SgsE_C-EGFP, SgsE_G-EGFP, SgsE_M-EGFP, and MBP-EGFP (Fig. 4). Single unfolding peaks were observed for SgsE_C and EGFP with melting temperatures (Tm) of 70.89 ± 0.03°C and 81.25 ± 0.03°C, respectively.
Fig. 4.
Differential scanning calorimetry measurements.
DSC signals were fitted with a non-2-state thermal unfolding model (red lines). Tm1 indicates thermal unfolding of either SgsE_C, SgsE_G, or MBP moieties, and Tm2 indicates thermal unfolding of the EGFP moiety, when applicable.
Two thermal unfolding peaks were observed for the chimeras SgsE_C-EGFP, SgsE_M-EGFP, and MBP-EGFP. In these thermograms, the first peak corresponds to unfolding of either the SgsE_C, SgsE_M, or MBP moiety, and the measured Tm values were 70.94 ± 0.01°C, 71.38 ± 0.03°C, and 64.30 ± 0.01°C, respectively. The second peak corresponds to the fused EGFP moiety with Tm values of 80.76 ± 0.04°C, 80.49 ± 0.06°C, and 81.38 ± 0.03°C, respectively.
SgsE_M-EGFP exhibited only a single peak with a Tm value of 80.06 ± 0.03°C, corresponding to the EGFP fusion part, whereas no signal for the SgsE_M moiety was detected. We interpreted this observation as the presence of an unstable and hardly folded state of this highly truncated SgsE form, in accordance with the above protein unfolding and refolding results.
Overall, melting temperatures of the individual fusion moieties appeared to be independent of each other. EGFP exhibited a Tm value of approximately 81°C regardless of its fusion partner, with the obtained value corresponding well to literature [17, 22]. The Tm value for fused MBP was 64°C, which is in good agreement with the literature [19]. Sole SgsE_C as well as fused SgsE_C and fused SgsE_G showed nearly identical Tm values, close to 71°C.
Discussion
Proteins are the workhorses of living cells and with the rise of biotechnology they are used in a vast range of medical and technical applications. However, each protein has evolved in a specific environment and usually requires specific conditions for correct folding and for fulfilling its intended function. Fusion of recombinantly produced proteins to highly soluble proteins is a possibility to increase the solubility of the target protein and prevent aggregation [7]; however, correct folding is not necessarily guaranteed. A more direct way of stabilizing a target protein might be desired to enable initial correct folding or to protect a protein in non-ideal environments.
The effect that SgsE_C was shown to exert on EGFP in the course of this study fulfills these tasks, whereas more truncated forms of SgsE did not. Folding of EGFP was enabled in an unsuitable environment when the fusion protein SgsE_C-EGFP was expressed with an N-terminal PelB signal sequence mediating Sec-dependent export of the unfolded chimera to the periplasmic space of E. coli BL21 cells. Periplasmic fluorescence was observed by fluorescence microscopy, whereas control cells exporting MBP-EGFP or EGFP did not show fluorescence. The inherent incapability of EGFP to fold correctly in the periplasm was thus overcome by fusion to SgsE_C.
Under more favorable in vitro conditions, refolding of EGFP fused to SgsE_C was observed to reach a higher extent of green fluorescence recovery compared with EGFP alone or the other EGFP fusion constructs investigated, corroborating the folding-enhancing effect.
The time-dependent decrease of fluorescence in buffered 3 M GdmCl solution was shown to be far less pronounced for the SgsE_C-EGFP chimera compared with EGFP alone and SgsE_G-EGFP, pointing to a protective influence of SgsE_C.
The nature of the observed effect is not likely to be triggered by increased solubility. Although SgsE_C might keep fused EGFP proteins at a distance, considering its size and assumed lengthy shape, a much stronger effect would be expected from MBP, which is known to be a potent solubility enhancer for GFP and other proteins [18]. Moreover, all measurements were performed at low concentrations, limiting the effects of GFP aggregation [9].
Since fusion to SgsE_C did not alter the thermal stability of the EGFP domain nor change the unfolding equilibria induced by chemical denaturation, the observed effect appears not to result from changed free folding energy. The most likely explanation is rather found in altered folding kinetics, which we directly observed in terms of different apparent rate constants.
The folding energy landscape of green fluorescent proteins was shown in different studies to be of complex nature, exhibiting intermediate states and alternative routes [5, 6, 12, 16, 20]. It was argued that high energy barriers exist for both folding and unfolding of GFP [12]. The effect from SgsE_C might therefore result from a reduction of the degrees of freedom for folding of EGFP in the fused state. EGFP closely attached to SgsE_C might not be able to explore all possible folding pathways and, thus, more likely follows a route leading to an active form. S-Layer proteins are in general stably folding themselves, implicating that SgsE_C will neither hamper the folding of EGFP, nor will it be distracted itself by a non-folded fusion partner. Related to its considerable size of 82.8 kDa, SgsE_C might in addition alter the environment encountered by EGFP in terms of hydrophobicity and net charge. It might also attenuate the effects of periplasmic constituents or denaturing agents that might impair folding of EGFP.
The observed stabilizing effect of SgsE_C might at least be in part specifically enacted on the EGFP moiety. However, in a previous work, it was already shown to significantly prolong the shelf-life of fused glucose-1-phosphate thymidylyltransferase RmlA [21], pointing to a more general stabilizing property of SgsE_C. Both the inherent capability of SgsE_C to self-assemble and the functions of fused proteins, such as the pH dependence of EGFP activity and the enzymatic function of fused RmlA, remained unchanged in SgsE_C-fusion proteins [15, 21]. Taken together, our results demonstrate a remarkable aspect of the S-layer protein from G. stearothermophilus NRS 2004/3a on the stabilization of two fused model proteins that supports the use of SgsE in fusion protein applications. The question of whether the SgsE S-layer acts in stabilizing or protecting surface-located enzymes in the native organism, which would indicate a novel biological function for S-layer proteins, remains to be addressed.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Austrian Science Fund FWF, projects P19047-B12 and P21954-B20 (to C.S.), P22791-B11 (to P.M.), the Christian Doppler Laboratory for Antibody Engineering (to C.O. and G.S), the PhD programme “BioToP - Biomolecular Technology of Proteins” (FWF project W1224), and the Programmausschreibung Forschungsinfrastruktur 4 of the Austrian Federal Ministry of Science and Research.
Footnotes
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
REFERENCES
- 1.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi SI, Ryu K, Seong BL. RNA-mediated chaperone type for de novo protein folding. RNA Biol. 2009;6:21–24. doi: 10.4161/rna.6.1.7441. [DOI] [PubMed] [Google Scholar]
- 3.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 4.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 5.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enoki S, Saeki K, Maki K, Kuwajima K. Acid denaturation and refolding of green fluorescent protein. Biochemistry. 2004;43:14238–14248. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 7.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000;182:4068–4076. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 10.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; New York: 2005. pp. 571–607. [Google Scholar]
- 11.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 12.Huang JR, Craggs TD, Christodoulou J, Jackson SE. Stable intermediate states and high energy barriers in the unfolding of GFP. J. Mol. Biol. 2007;370:356–371. doi: 10.1016/j.jmb.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Ikura K, Kokubu T, Natsuka S, Ichikawa A, Adachi M, Nishihara K, Yanagi H, Utsumi S. Co-overexpression of folding modulators improves the solubility of the recombinant guinea pig liver transglutaminase expressed in Escherichia coli. Prep. Biochem. Biotechnol. 2002;32:189–205. doi: 10.1081/PB-120004130. [DOI] [PubMed] [Google Scholar]
- 14.Ilk N, Egelseer EM, Sleytr UB. S-Layer fusion proteins - construction principles and applications. Curr. Opin. Biotechnol. 2011;22:824–831. doi: 10.1016/j.copbio.2011.05.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kainz B, Steiner K, Möller M, Pum D, Schäffer C, Sleytr UB, Toca-Herrera JL. Absorption, steady-state fluorescence, fluorescence lifetime, and 2D self-assembly properties of engineered fluorescent S-layer fusion proteins of Geobacillus stearothermophilus NRS 2004/3a. Biomacromolecules. 2010;11:207–214. doi: 10.1021/bm901071b. [DOI] [PubMed] [Google Scholar]
- 16.Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proc. Natl. Acad. Sci. USA. 2007;104:20268–20273. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Málnási-Csizmadia A, Somogyi B, Lörinczy D. Thermal stability of chemically denatured green fluorescent protein (GFP): A preliminary study. Thermochim. Acta. 2004;410:161–163. [Google Scholar]
- 18.Nallamsetty S, Waugh DS. Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr. Purif. 2006;45:175–182. doi: 10.1016/j.pep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Novokhatny V, Ingham K. Thermodynamics of maltose binding protein unfolding. Protein Sci. 1997;6:141–146. doi: 10.1002/pro.5560060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J. Biol. Chem. 2006;281:40010–40014. doi: 10.1074/jbc.M609890200. [DOI] [PubMed] [Google Scholar]
- 21.Schäffer C, Novotny R, Küpcü S, Zayni S, Scheberl A, Friedmann J, et al. Novel biocatalysts based on S-layer self-assembly of Geobacillus stearothermophilus NRS 2004/3a: A nanobiotechnological approach. Small. 2007;3:1549–1559. doi: 10.1002/smll.200700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheyhing CH, Meersman F, Ehrmann MA, Heremans K, Vogel RF. Temperature–pressure stability of green fluorescent protein: A Fourier transform infrared spectroscopy study. Biopolymers. 2002;65:244–253. doi: 10.1002/bip.10237. [DOI] [PubMed] [Google Scholar]
- 23.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 24.Sleytr UB, Messner P, Pum D, Sára M. Crystalline bacterial cell surface layers. Mol. Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 25.Steiner K, Hanreich A, Kainz B, Hitchen PG, Dell A, Messner P, Schäffer C. Recombinant glycans on an S-layer self-assembly protein: A new dimension for nanopatterned biomaterials. Small. 2008;4:1728–1740. doi: 10.1002/smll.200701215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 27.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 28.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 29.Xie JB, Zhou JM. Trigger factor assisted folding of green fluorescent protein. Biochemistry. 2008;47:348–357. doi: 10.1021/bi7011838. [DOI] [PubMed] [Google Scholar]
- 30.Zarschler K, Janesch B, Kainz B, Ristl R, Messner P, Schäffer C. Cell surface display of chimeric glycoproteins via the S-layer of Paenibacillus alvei. Carbohydr. Res. 2010;345:1422–1431. doi: 10.1016/j.carres.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.